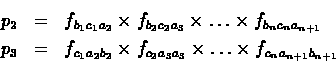Next: Detecting Promoter Regions
Up: Gene Finding
Previous: Finding Long ORF's
Compositional Differences
A more informative method
to determine coding regions, takes advantage of the frequencies in
which the various codons occur in coding regions. For example, the
amino acids Leucine, Alanine and Tryptophan are coded by 6,4 and 1
different codons respectively. In a translation of a uniformly
random DNA sequence, these amino acids should occur in the ratio
6:4:1, but in a protein they occur in a different ratio -
6.9:6.5:1. Therefore coding DNA is not random. Another example of
the non-uniformness of coding DNA is the fact that A or T occurs
in the 3rd position of a codon 90% of the time, while G or C
occur only 10% of the time (these statistics are different for
different species).
Assume that
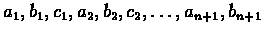 is a
coding sequence of bases with an unknown reading frame. Let
fabc denote the frequency with which the codon abc appear
in a coding region. Under the assumption that successive codons
are chosen independently at random according to the frequencies
fabc, the probability of observing the sequence of n codons
appearing in the reading frame starting with a1b1c1 is
is a
coding sequence of bases with an unknown reading frame. Let
fabc denote the frequency with which the codon abc appear
in a coding region. Under the assumption that successive codons
are chosen independently at random according to the frequencies
fabc, the probability of observing the sequence of n codons
appearing in the reading frame starting with a1b1c1 is
Similarly, the probability of observing the n
codons in the second and third coding frames are :
The probability Pi which is the probability of the ith
reading frame being the translated one (assuming the region is
coding) can be calculated as follows:
Figure 7.6:
Application of
codon frequency method to bases 1 to 10,000 of the liverwort
Marchantia chloroplast genome. The horizontal scale marks every
100th base, and the bars above indicate the extent of known
protein coding segments. The three boxes above contain plots of
the probability that each of the three reading frames is coding
for a protein. The short vertical lines that bisect the mid-height
of each box mark the positions of the stop codons in the
corresponding reading frames. The vertical scale within each box
is of
log(P/1-P), so that, for example, 4 points up the scale
from the mid-height corresponds to 99.99% probability of the
current region being coding.
|
The
above computation can be used in a search algorithm as follows:
Slide a window of size n along the sequence, and compute Pi
for each start position of the window. The Codon Preference
program, which is part of the GCG library, implements this
method.
Figure 7.6 shows the plot of
log(P/1-P), which is the log likelihood, for the three reading
frames. It shows that the value of
log(P/1-P) is usually above
4 ( 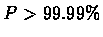 ) in regions where the DNA sequence codes for a protein in
that reading frame.
This method depends on the accuracy of the codon frequency
statistics of already found genes. The algorithm will also have
difficulty in detecting horizontal gene transfer and other causes of
heterogeneity.
) in regions where the DNA sequence codes for a protein in
that reading frame.
This method depends on the accuracy of the codon frequency
statistics of already found genes. The algorithm will also have
difficulty in detecting horizontal gene transfer and other causes of
heterogeneity.
Figure:
Periodicity of T: for each gap size n,
the number of occurrences of the pattern T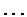 T, with n bases
between the two T's, was counted. Plotted is the present
difference of that number from the number of such pair expected if
the bases occurred at random. Since the T's occur preferentially
at the second codon position, in coding regions, the present
difference at n=2,5,8,... is noticeably
large.
T, with n bases
between the two T's, was counted. Plotted is the present
difference of that number from the number of such pair expected if
the bases occurred at random. Since the T's occur preferentially
at the second codon position, in coding regions, the present
difference at n=2,5,8,... is noticeably
large.
|
Various other methods have
also been tried. One variation of the codon frequency method is to
use 6-tuple frequencies, which are more informative than 3-tuple
(codon) frequencies. Another approach (which is illustrated in
figure 7.7) considers the periodicity of
certain bases in protein coding regions, whose statistical
behaviour distinguishes them from non coding regions.



Next: Detecting Promoter Regions
Up: Gene Finding
Previous: Finding Long ORF's
Itshack Pe`er
1999-02-03