- 1.
- For each species, compute
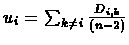 .
.
- 2.
- Choose the i and j for which Di,j - ui - uj is smallest.
- 3.
- Join clusters i and j to a new cluster - (ij), with a corresponding
node in T. Calculate the branch lengths from i and j to the new node as:
- 4.
- Compute the distances between the new cluster and each other cluster:
- 5.
- Delete clusters i and j from the tables, and replace them by (ij).
- 6.
- If more than two nodes (clusters) remain, go back to 1. Otherwise, connect the two remaining nodes by a branch of length Di,j.