


Next: Identification of Gene Regulatory
Up: Boolean Network Model
Previous: Boolean Network Model
In order to infer a boolean genetic network, a population of cells
containing a target genetic network is monitored in the steady
state over a series of M experimental perturbations. In each
perturbation Pm (
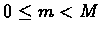 )
any number of nodes may be
forced either to a low or a high level. Genes may be perturbed
through laboratory methods for gene deletion or overexpression,
while stimuli are perturbed by altering the environment outside
the cell. The observed steady-state expression levels for all
genes and stimuli over all experimental perturbations or over time
are represented by the expression matrix E.
Figure 14.8 shows an expression matrix generated by
several illustrative perturbations. Rows of E represent perturbation
conditions while columns represent node values in each
steady-state condition, such that matrix entry Eij is the
expression level of node Xi in the presence of perturbation
Pj. The symbols "+" and "-" are used to show that a node has
been forced to a high or low value, respectively.
)
any number of nodes may be
forced either to a low or a high level. Genes may be perturbed
through laboratory methods for gene deletion or overexpression,
while stimuli are perturbed by altering the environment outside
the cell. The observed steady-state expression levels for all
genes and stimuli over all experimental perturbations or over time
are represented by the expression matrix E.
Figure 14.8 shows an expression matrix generated by
several illustrative perturbations. Rows of E represent perturbation
conditions while columns represent node values in each
steady-state condition, such that matrix entry Eij is the
expression level of node Xi in the presence of perturbation
Pj. The symbols "+" and "-" are used to show that a node has
been forced to a high or low value, respectively.
Figure:
Expression Matrix - adapted from [5]

|
Peer Itsik
2001-03-04
