


Next: Expression Matrix
Up: Gene Networks
Previous: Gene Network Models
Boolean Network Model
According to this model the network is
represented as an oriented graph
G = (V, F), whose nodes V
represent element of the network, and F defines a topology of
edges between the nodes and a set of boolean functions.
A node may represent either a gene or a biological stimulus, where a stimulus is any relevant
physical or chemical factor which influences the network and is itself not a gene or
gene product. A node has an associated steady-state expression level xv ,
representing the amount of gene product (in the case of a gene) or the amount of
stimulus present in the cell. This level is approximated as high or low and
represented by the binary value 1 or 0, respectively.
Network behavior over time is modelled as a sequence of discrete
synchronous steps. The set
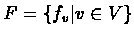 of boolean
functions assigned to the nodes defines the value of a node on the
next step depending on values of other nodes, which influence it.
The functions fv are uniquely defined using truth tables. An
edge directed from one node to another represents the influence of
the first gene or stimulus on that of the second, so that the
expression level of a node v is a Boolean function fv of the
levels of the nodes in the network which connect (have a directed
edge) to v. We sometimes confuse the network with its undirected
underlying graph G(V,E).
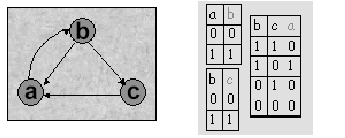
The following figures 14.6 give an example of a simple boolean
network and associated truth tables. This example shows a network of three
nodes - a, b and c. As one can see, expression of c directly
depends on expression of b, which directly depends on a. However, influence
of b and c on a is more complex. For example, high level of expression
of both b and c leads to inhibition of a.
of boolean
functions assigned to the nodes defines the value of a node on the
next step depending on values of other nodes, which influence it.
The functions fv are uniquely defined using truth tables. An
edge directed from one node to another represents the influence of
the first gene or stimulus on that of the second, so that the
expression level of a node v is a Boolean function fv of the
levels of the nodes in the network which connect (have a directed
edge) to v. We sometimes confuse the network with its undirected
underlying graph G(V,E).
The following figures 14.6 give an example of a simple boolean
network and associated truth tables. This example shows a network of three
nodes - a, b and c. As one can see, expression of c directly
depends on expression of b, which directly depends on a. However, influence
of b and c on a is more complex. For example, high level of expression
of both b and c leads to inhibition of a.
Figure:
Sample boolean network - adapted from [8]
|
The assignment of values to nodes fully describes the state of
the model at any given time. The change of model state over time
is fully defined by the functions in F. Initial assignment of
values uniquely defines the model state at the next step, and,
consequently, on all the future steps. Thus, the network evolution
is realized as a sequence of consecutive states, and it is
uniquely defined by the initial state. Such a sequence of states
is called trajectory.
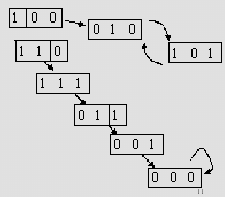
Figure 14.7 shows two such trajectories for our
sample network. Since the number of possible states is finite, all
trajectories eventually end up in single steady state, or
a cycle of steady states.
Figure:
States trajectories - adapted from [8]
|
- An attractor of a trajectory is a single steady state, or a cycle
at the end of the trajectory.
- The basin of attraction for a specific attractor is a set of all trajectories leading
to it.
The network in our example has two attractors - one is the steady
state (0,0,0), and the other is a cycle
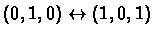 .
States in gene networks are often characterized by
stability - "slight" changes in value of a few nodes do
not change the attractor. Biological systems are often
redundant to ensure that the system stays stable and
retains its function even in presence of local anomalies. For
example, there may be two proteins, or even two different networks
with the same function, which backup each other.
.
States in gene networks are often characterized by
stability - "slight" changes in value of a few nodes do
not change the attractor. Biological systems are often
redundant to ensure that the system stays stable and
retains its function even in presence of local anomalies. For
example, there may be two proteins, or even two different networks
with the same function, which backup each other.



Next: Expression Matrix
Up: Gene Networks
Previous: Gene Network Models
Peer Itsik
2001-03-04
