


Next: Multi-Condition Gene Expression Data
Up: Clustering Using BioClust
Previous: Synthetic Data
The gene expression data used in this experiment is from [8]. In this paper the authors study the relationship among expression patterns of genes involved in the rat Central Nervous System (CNS).
Gene expression patterns were measured for 112 genes along nine different development time points. The experiments here are the time points. The gene expression data for each gene was augmented with derivative values to enhance the similarity for closely parallel but offset expression patterns resulting in
expression matrix. The similarity matrix was obtained using Euclidean distance.
Eight clusters were obtained. Since partitioning to clusters is known from [8] this experiment was done mainly for validation of algorithm.
Figures 12.5 and 12.6 present the clustering results. Note that all clusters, perhaps with exception of cluster #1, manifest clear and distinct expression patterns. Moreover, the agreement with the prior biological classification is quite good.
Figure:
Applying the algorithm to temporal gene expression data [8]. The solution generated by the algorithm is compared to the prior classification. For each cluster (x-axis), bars composition in terms of biologically defined families. The height of each bar (y-axis) represents the number of genes of a specific cluster family. Most clusters contain predominantly genes from one or two families.
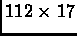
|
Figure:
Applying the algorithm to temporal gene expression data [8]. Each graph presents expression patterns of genes in a specific cluster. The x-axis represents time, while the y-axis represents normalized expression level.
|
|



Next: Multi-Condition Gene Expression Data
Up: Clustering Using BioClust
Previous: Synthetic Data
Peer Itsik
2001-02-01