


Next: Temporal Gene Expression Data
Up: Clustering Using BioClust
Previous: Clustering Using BioClust
BioClust is tested on the members of
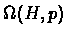 .
The simulation procedure is as follows (please refer to figure 12.3 for visualization of the simulation procedure):
.
The simulation procedure is as follows (please refer to figure 12.3 for visualization of the simulation procedure):
- Generate G from H by independently removing each edge in H with probability p and adding each edge not in H with probability p;
- Randomly permute the order of vertices in G and run BioClust with affinity threshold
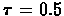 ;
;
- Compare BioClust's output to the original clique graph H;
Figure:
A visualization of the simulation procedure. A: The adjacency matrix of the original clique graph H before introduction of errors. Position (i,j) is white if
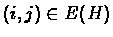 ,
that is, if i and j belong to the same cluster. B: The same matrix after introduction of errors. Note that the cluster structure is still visible for all but the smallest clusters. C: The same as B but vertex order is randomly permuted. This is the actual input to the algorithm. D: Matrix C reordered according to solution produced by the algorithm. With the exception of perhaps the smallest clusters, the essential cluster structure is reconstructed.
,
that is, if i and j belong to the same cluster. B: The same matrix after introduction of errors. Note that the cluster structure is still visible for all but the smallest clusters. C: The same as B but vertex order is randomly permuted. This is the actual input to the algorithm. D: Matrix C reordered according to solution produced by the algorithm. With the exception of perhaps the smallest clusters, the essential cluster structure is reconstructed.
|
|
There are several comparison criteria, which can be used to compare the algorithm's output to the original clique graph. The matching coefficient is defined by
 that is total number of matching entries divided by total number of entries. The Jaccard coefficient is defined by
that is total number of matching entries divided by total number of entries. The Jaccard coefficient is defined by
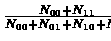 ,
which is similar to the matching coefficient, only with N00 the number of entries which are zero in both matrices removed. In sparse graphs N00 will be a dominant factor, thus Jaccard coefficient is more sensitive when dealing with sparse graphs.
,
which is similar to the matching coefficient, only with N00 the number of entries which are zero in both matrices removed. In sparse graphs N00 will be a dominant factor, thus Jaccard coefficient is more sensitive when dealing with sparse graphs.
Table 12.1:
Performance of the BioClust algorithm for different values of p. Mean values of matching coefficient and Jaccard coefficient are given.
| |
| cluster structure |
n |
p |
matching coeff. |
Jaccard coeff. |
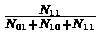 |
500 |
0.2 |
1.0 |
1.0 |
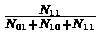 |
500 |
0.3 |
0.999 |
0.995 |
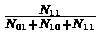 |
500 |
0.4 |
0.939 |
0.775 |
|
Table 12.1 presents results of simulation for different values of contamination error p. The values of the matching coefficient and the Jaccard coefficient are presented. It can be seen that the Jaccard coefficient is more sensitive. One can also observe the effect of p on the performance of the algorithm.
Figure 12.4 presents results of simulation for different values of n and p. It can be seen that the properties of the theoretical algorithm are preserved in its practical implementation. We get better performance when the number of clustered entities increases.



Next: Temporal Gene Expression Data
Up: Clustering Using BioClust
Previous: Clustering Using BioClust
Peer Itsik
2001-02-01
 that is total number of matching entries divided by total number of entries. The Jaccard coefficient is defined by
that is total number of matching entries divided by total number of entries. The Jaccard coefficient is defined by
![]() ,
which is similar to the matching coefficient, only with N00 the number of entries which are zero in both matrices removed. In sparse graphs N00 will be a dominant factor, thus Jaccard coefficient is more sensitive when dealing with sparse graphs.
,
which is similar to the matching coefficient, only with N00 the number of entries which are zero in both matrices removed. In sparse graphs N00 will be a dominant factor, thus Jaccard coefficient is more sensitive when dealing with sparse graphs.