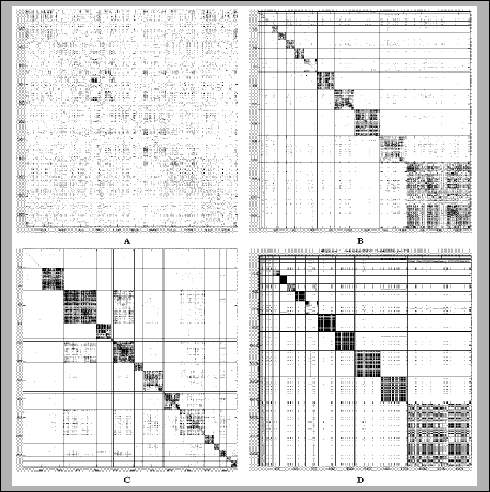
| Cluster |
Cluster size |
Gene name |
| T18 |
709 |
Ef1 alpha |
| T17 |
285 |
clone 190B1 |
| T16 |
284 |
Cytochr c oxi |
| T15 |
213 |
tubulin beta |
| T14 |
187 |
40SRibo protS6 |
| T13 |
146 |
40SRibo protS3 |
| T12 |
108 |
40SRibo protS4 |
| T11 |
91 |
GAPDH |
| T10 |
86 |
60SRibo protL4 |
| T9 |
67 |
Ef1 beta |
| T8 |
43 |
Human calmodulin |
| T7 |
39 |
heat shock cogKD71 |
| T6 |
32 |
heat shock cogKD90 |
| T5 |
14 |
Human TNF recep |
| T4 |
12 |
Human AEBP1 |
| T3 |
10 |
clone 244D14 |
| T2 |
2 |
clone 241F17 |
| T1 |
1 |
Human anion ch |
|