Next: Clustering Real cDNA Data
Up: cDNA Clustering
Previous: Assessing Clustering Quality
Intensive tests of the algorithm on simulated data were performed. The
simulation process computes artifical gene fingerprints (hybridized oligos) for
each participating gene. For each gene and a given probe, the precise
locations along the gene are generated in a realistic manner. Then, truncated
clones of each gene are generated. Each clone inherits the probe fingerprints
and
their locations from its original gene (just the fingerprints with locations
within the clone boundaries are inherited).
Finally, each copy is incorporated with
false positive and false negative errors, again, realistically. If we denote
the total number of oligos by p and the total number of clones by N, then
the result of the simulation is an
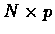 hybridization matrix H,
where Hij=1 if clone i hybridized with oligo j, and Hij=0
otherwise.
The simulation results are summarized in figure 11.10.
A comparison of the Minkowski scores is given in
figure 11.11.
hybridization matrix H,
where Hij=1 if clone i hybridized with oligo j, and Hij=0
otherwise.
The simulation results are summarized in figure 11.10.
A comparison of the Minkowski scores is given in
figure 11.11.
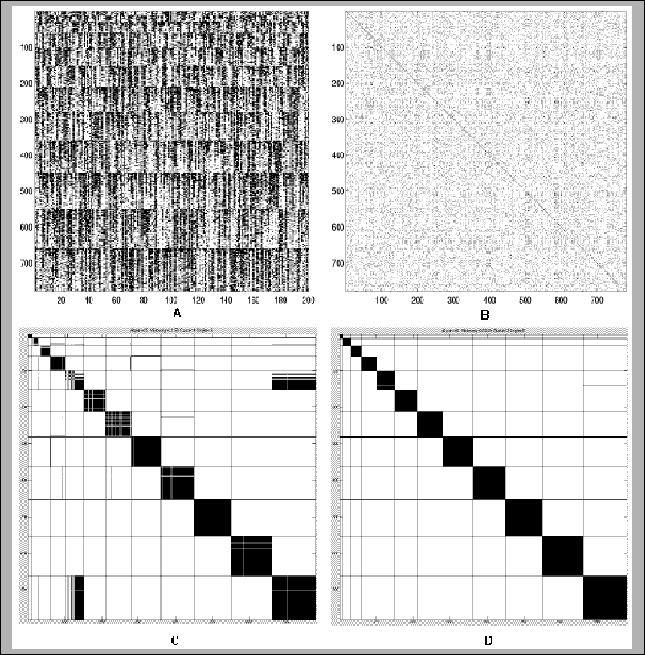
Figure 11.10:
Examples of results of HCS
and Greedy clustering algorithms in high noise simulation. The fingerprint
data consisted of 780 cDNAs from 12 genes, in clusters of sizes 10,20,...,120.
The number of oligos is 200. The expected rate of false positive
hybridizations is 25%. The expected false negative hybridization rate is
40%. A: The hybridization fingerprints matrix H. Each of the 780 rows is a
fingerprint vector of one cDNA. White denotes positive hybridization. B: The
binarized similarity matrix. Position i,j is black iff Sij>50. Matrix
coordinates are scrambled, as in realistic scenarios. C: Clustering solution
generated by the greedy algorithm. Minkowski score is 1.32. cDNAs from the
same true cluster appear consecutively, and the black lines are the borders
between the different clusters. Position i,j is black if the solution puts
cDNAs i and j in the same cluster. D: Clustering solution generated by the
HCS algorithm. Minkowski score is 0.209.
|
|
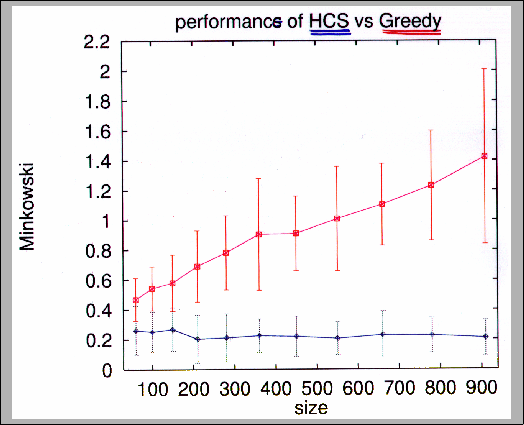
Figure 11.11:
Performance comparison of HCS (squares) and Greedy (diamonds)
algorithms on simulation data (using Minkowski score).
|
|



Next: Clustering Real cDNA Data
Up: cDNA Clustering
Previous: Assessing Clustering Quality
Peer Itsik
2001-01-31