 where
N(t) = number of events until time t
where
N(t) = number of events until time t
| (2) |
As as consequence, distribution of the number of events in an interval is stationary, i.e. depends only on the length of the interval. The expected number of events in an interval of length t is given by
 are distributed uniformly and
independently in [0,t].
Assume clone length L, genome length G, and choose N clones
at random. What is the expected fraction of the genome covered by
clones?
For a random point b, and an arbitrary clone C the probability
of the point b being included in the clone c is given by:
are distributed uniformly and
independently in [0,t].
Assume clone length L, genome length G, and choose N clones
at random. What is the expected fraction of the genome covered by
clones?
For a random point b, and an arbitrary clone C the probability
of the point b being included in the clone c is given by:
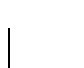 and
and 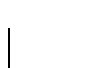 .
The fraction
.
The fraction