To align the string
![]() against a profile
against a profile
![]() of length L, we will use a variant of the Viterbi
algorithm. For each
of length L, we will use a variant of the Viterbi
algorithm. For each
![]() and
and
![]() we use the
following definitions:
we use the
following definitions:
The initial value of the special begin state is:
| vbegin(0) = 0 | (44) |
To calculate the values of vMj(i), vIj(i) and vDj(i) we use the same technique as in the Viterbi algorithm. There are however two major differences:
The three predecessors of the match state Mj are the three states
of the previous layer, j-1:
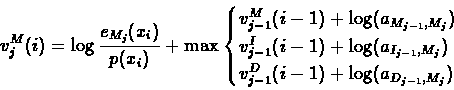 |
(45) |
The three predecessors of the insertion state Ij are the
three states of the same layer, j:
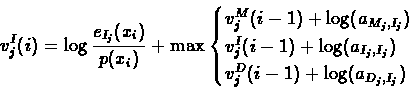 |
(46) |
The three predecessors of the deletion state Dj are the three
states of the layer j-1. Since Dj is a silent state, we
should not consider the emission likelihood score for xi in
this case:
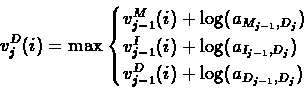 |
(47) |
We conclude by calculating the optimal score:
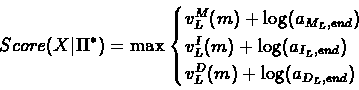 |
(48) |
Complexity: We have to calculate
![]() values,
while calculating each value takes O(1) operations (since we
only need to consider the scores of at most three predecessors). We
therefore need
values,
while calculating each value takes O(1) operations (since we
only need to consider the scores of at most three predecessors). We
therefore need
![]() time and
time and
![]() space.
space.
We can use a similar approach for the problem of local alignment of a sequence versus a profile HMM. This is achieved by adding four additional states (the lightly shaded states in figure 6.6) corresponding to the alignment of a sub-string of X to a part of the profile.