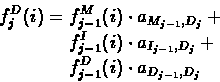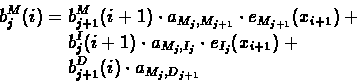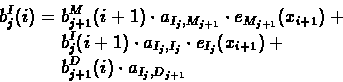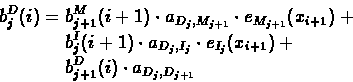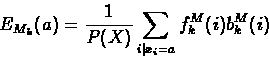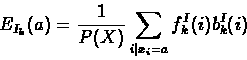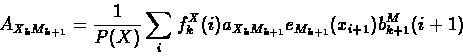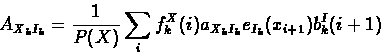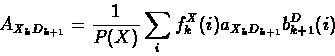Next: Multiple Alignment with Profile
Up: Profile Alignment
Previous: Aligning Sequences to a
In the previous section we modeled the problem of aligning a
string to a profile. As with general HMMs, the main problem is to
assign meaningful values to the transition and emission
probabilities to a profile HMM. It is possible to use the
Baum-Welch algorithm for training the model probabilities, but it
first has to be shown how to compute the forward and backward
probabilities needed for the algorithm.
Given a string
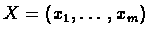 we define:
we define:
- The forward probabilities:
- The backward probabilities:
Computing the Forward Probabilities:
- 1.
- Initialization:
- 2.
- Recursion:
![\begin{displaymath}\begin{split}
f_{j}^{M}(i) = e_{M_{j}}(x_{i}) \, \cdot \,
[...
...}+ \\
&f^{D}_{j-1}(i-1)\cdot a_{D_{j-1},M_{j}}]
\end{split} \end{displaymath}](img123.gif) |
(56) |
![\begin{displaymath}\begin{split}
f^{I}_{j}(i) = e_{I_{j}}(x_{i}) \, \cdot \,
[...
...I_{j}}+\\
&f^{D}_{j}(i-1)\cdot a_{D_{j},I_{j}}]
\end{split} \end{displaymath}](img124.gif) |
(57) |
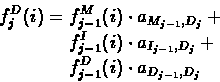 |
(58) |
Computing the Backward Probabilities:
- 1.
- Initialization:
- 2.
- Recursion:
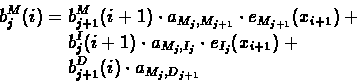 |
(62) |
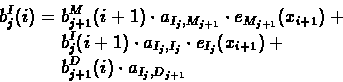 |
(63) |
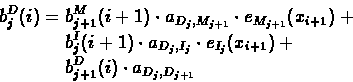 |
(64) |
The forward and backward variables can then be combined to re-estimate emission and transition probability parameters as follows:
Baum-Welch re-estimation equations fo profile HMMs:
- 1.
- Expected emission counts from sequence X:
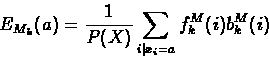 |
(65) |
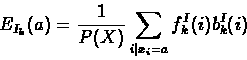 |
(66) |
- 2.
- Expected transition counts from sequence x:
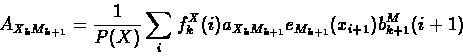 |
(67) |
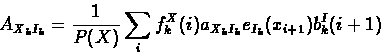 |
(68) |
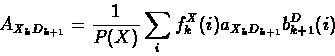 |
(69) |



Next: Multiple Alignment with Profile
Up: Profile Alignment
Previous: Aligning Sequences to a
Peer Itsik
2000-12-19
![]() we define:
we define:
![\begin{displaymath}\begin{split}
f_{j}^{M}(i) = e_{M_{j}}(x_{i}) \, \cdot \,
[...
...}+ \\
&f^{D}_{j-1}(i-1)\cdot a_{D_{j-1},M_{j}}]
\end{split} \end{displaymath}](img123.gif)
![\begin{displaymath}\begin{split}
f^{I}_{j}(i) = e_{I_{j}}(x_{i}) \, \cdot \,
[...
...I_{j}}+\\
&f^{D}_{j}(i-1)\cdot a_{D_{j},I_{j}}]
\end{split} \end{displaymath}](img124.gif)