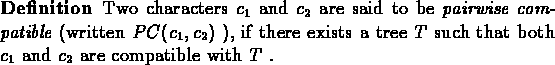
The more interesting problem here is of course that of ``large compatibility'' - finding the best phylogeny given only the data matrix M. We shall tackle this problem through the notions of pairwise compatibility and joint compatibility.
We will present two theorems. The first, by Wilson [16], identifies pairwise compatible characters:
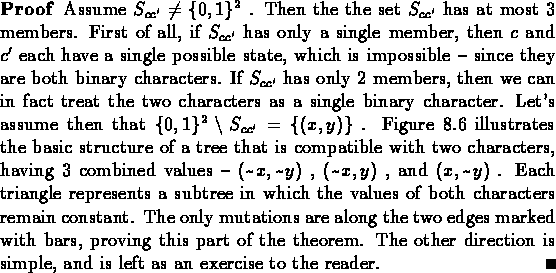
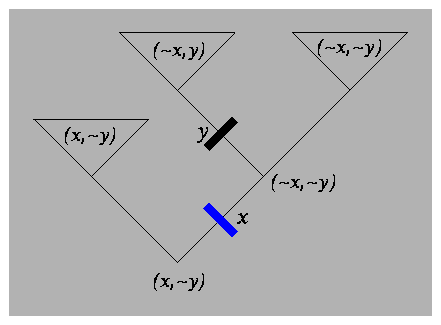 |
The next, somewhat surprising, theorem by Estabrook et al. [7] identifies jointly compatible sets of binary characters:
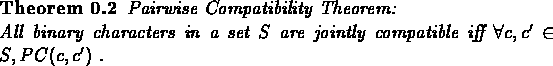
Note that the theorem does not hold for characters that are not binary. We will not present a proof for this theorem.
So the problem of ``large compatibility'' is reduced to the problem of finding the largest jointly compatible set of characters, which amounts to finding a maximum clique in the pairwise-compatibility graph, defined as:
This seems to be of no great help, because as we know, finding a maximum clique in a graph is an NP-hard problem. However, there are algorithms, such as Bron and Kerbosch's [1] Branch-and-Bound clique-finding algorithm, which seem to work very well with biological data. All in all, compatibility methods usually run faster than parsimony methods for the same data.
Perfect Phylogeny
The problem of perfect phylogeny (also called full compatibility) is to decide if all the characters are jointly compatible. For binary characters the problem is easy: it is solved by checking if the pairwise-compatibility graph is a complete graph. For non-binary characters the problem is NP-hard (it relates to some properties of chordal graphs).