| Elements |
Problem |
Compared to |
Improvement |
Time(min) |
| 517 |
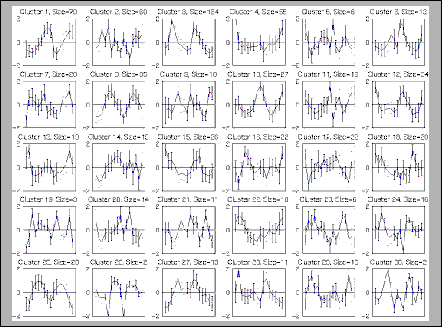
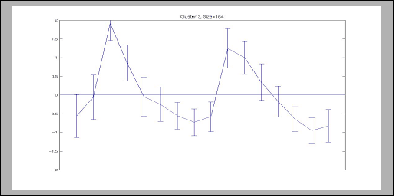
Gene Expression Fibroblasts |
Cluster [5] |
Yes |
0.5 |
| 826 |
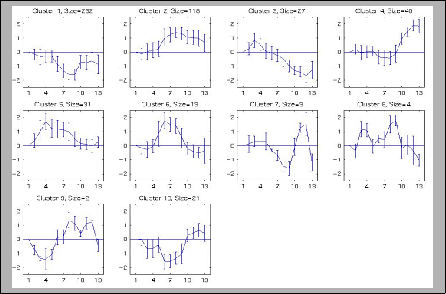
Gene Expression Yeast cell cycle |
GeneCluster [23] |
Yes |
0.2 |
| 2,329 |
cDNA OFP Blood Monocytes |
HCS [7] |
Yes |
0.8 |
| 20,275 |
cDNA OFP Sea urchin eggs |
K-Means [8] |
Yes |
32.5 |
| 72,623 |
Protein similarity |
ProtoMap [24] |
Minor |
53 |
| 117,835 |
Protein similarity |
SYSTERS [11] |
Yes |
126.3 |
|