


Next: Problem Statement
Up: Constructing Physical Maps from
Previous: Introduction
The Statistical
Model
We now present the work by Mayraz and Shamir [5] on the physical mapping from noisy data. The statistical model used for the above
mapping method assumes the following:
- 1.
- Clones are uniformly and independently distributed along the
target genome.
- 2.
- Clones are of equal length.
- 3.
- Probe occurrences along the genome are modeled by a
Poisson process.
- 4.
- The Poisson rate is identical for all probes.
- 5.
- The noise statistically behaves as follows:
- False positive errors - a Poisson process with parameter
.
- False negative errors independently occur with probability
for each hybridization
The hybridization scenario is shown by figure
9.8. The clones are the horizontal
lines. The random occurrences of a single nonunique probe are
marked by the dotted vertical lines. We denote by A the probe -
clone occurrence matrix:
Ai,j = k if probe j occurs k
times in clone i. The probe in this example occurs 3 times along
the 7 clones in this genomic region, so its column in the occurrence
matrix would be
(1, 1, 0, 0, 1, 2, 0). The probability of j
occurring k times in i is given by:
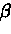 |
(7) |
We denote by B the probe clone hybridization matrix:
Bi,j
= 1 or
Bi,j = 0 depending on whether probe j hybridized
with clone i or not. The vector
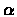 of the
hybridizations of clone i with all the probes is also called its
hybridization fingerprint. In case no noise is present
hybridization occurs iff there is at least one occurrence of the
probe. In this case the appropriate column of B would be
(1, 1,
0, 0, 1, 1, 0). Experimental noise can result in both false
positive hybridizations
(Bi,j = 1 when
Ai,j = 0), and
false negative hybridizations
(Bi,j = 0 when
Ai,j > 0).
of the
hybridizations of clone i with all the probes is also called its
hybridization fingerprint. In case no noise is present
hybridization occurs iff there is at least one occurrence of the
probe. In this case the appropriate column of B would be
(1, 1,
0, 0, 1, 1, 0). Experimental noise can result in both false
positive hybridizations
(Bi,j = 1 when
Ai,j = 0), and
false negative hybridizations
(Bi,j = 0 when
Ai,j > 0).
Hybridization fingerprints of intersecting clones are correlated.
This fact is used in order to estimate the clone pairs overlap.
Although noise reduces the correlation between fingerprints of
overlapping clones, Bayesian inference can still be used to
identify overlap, provided a sufficient number of probes is used.
It may also be the case that "soft decision" hybridization signals
are available. Such signals provide more information on probe
occurrences than binary signals do. This continuous signal value
does not directly correspond to the hybridization probability, and
we have chosen to assume a threshold is used to transform the
hybridization signal into a binary one. We therefore define the
hybridization matrix B to be a binary matrix, such that
Bi,j
= 1 if probe j has produced a positive hybridization signal
with clone i. The matrix B is the actual experimental data,
which is the input for the construction algorithm. The matrix
contains noise and no information on multiplicities. Using the
statistical model we can write the following equation:



Next: Problem Statement
Up: Constructing Physical Maps from
Previous: Introduction
Peer Itsik
2001-01-09