Next: UPGMA
Up: Distance Based Methods
Previous: Distance between DNA Sequences
Least Squares Methods
One of the more statistically justified methods to approximate a distance matrix
is the least squares approach. In this formulation we are giving, for each pair of species, the measurred distance
Di,j between them, and the weightwi,j that intuitively quuantifies the accuracy of this measure .Our goal is to find a tree T,
whose leaves are the n given species, and that predicts distances dijbetween the species, so that the following expression is minimized:
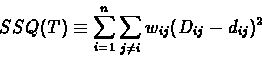 |
(6) |
The SSQ is a measure of the discrepancy between the observed
distances Di,j and the distances di,j predicted by T. The weights
wi,j are usually all 1, or
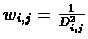 .
.

Again, a "small" version of this problam is formulated for a given tree, only trying to minimize SSQ by determining the branches length. In general, the "large" problam of finding the least squares tree is NP-complete [2].
We will discuss two polynomial heuristics - UPGMA and Neighbor-Joining. We have already
studied these algorithms in lecture #5, where we used them to iteratively add one additional
string to a growing multiple alignment, thus obtaining a progressive alignment.



Next: UPGMA
Up: Distance Based Methods
Previous: Distance between DNA Sequences
Peer Itsik
2001-01-01