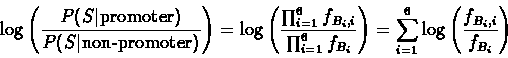Next: Promoter variation
Up: Detection of Promoter Regions
Previous: Positional Weight Matrix
Let fb denote the expected frequency of the base b in the
genome (the background frequency). We calculate the likelihood of
a given sequence being a TATA-box. For a sequence
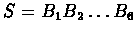 the likelihood of it being a TATA-box is:
the likelihood of it being a TATA-box is:
Similarly, the
likelihood of observing it, given it is a "non-promoter" is:
The log-likelihood ratio is
therefore:
This
model has the disadvantage that it doesn't exploit all of the
known information (i.e. dependencies between bases occurring in
the promoter regions etc.) The fBi are given in Figure
![[*]](/usr/local/lib/latex2html-98.1p1/icons.gif/cross_ref_motif.gif) .
.
Figure:
Positional
weight matrix for TATA box [].
|
|
Peer Itsik
2000-12-25