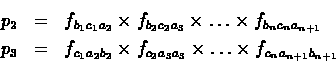Next: Detection of Promoter Regions
Up: Detection of Coding Regions
Previous: ORFs as Markov chains
In the model described above the probability of a codon occurrence
depends on the preceding codon. We now consider a simpler model in
which successive codons are independent. Let fabc denote the
frequency with which the codon abc occurs in a coding region.
Given a coding sequence
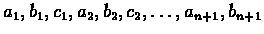 with an unknown reading frame, the probability
of observing the sequence of n codons appearing in the reading
frame starting with a1b1c1 is
with an unknown reading frame, the probability
of observing the sequence of n codons appearing in the reading
frame starting with a1b1c1 is
Similarly, the
probability of observing the n codons in the second and third
coding frames are:
Let Pi denote the probability of the ith reading frame being
the coding reading frame (assuming the region is coding). Pi
can be calculated as follows:
The above computation can be used in a search algorithm as
follows: Slide a window of size n along the sequence, and
compute Pi for each start position of the window. The
Codon Preference program, which is part of the GCG library,
implements this method.
Figure:
Results of
codon preference program [1].
|
|
Figure ![[*]](/usr/local/lib/latex2html-98.1p1/icons.gif/cross_ref_motif.gif) shows a the plot of
log(P/1-P), which is the log likelihood, for the three reading
frames. Each point represents the score for a 25 codon window
around it. The actual genes are plotted as rectangles at the
bottom. We can see that in the reading frame matching the upper
plot, the genes are clearly recognized.
shows a the plot of
log(P/1-P), which is the log likelihood, for the three reading
frames. Each point represents the score for a 25 codon window
around it. The actual genes are plotted as rectangles at the
bottom. We can see that in the reading frame matching the upper
plot, the genes are clearly recognized.
Figure:
Results of
codon preference program - 3rd position bias [1].
|
|
Figure ![[*]](/usr/local/lib/latex2html-98.1p1/icons.gif/cross_ref_motif.gif) shows the plot of a program
using only the 3rd position bias information. These methods
depend on the accuracy of the codon frequency statistics of
already found genes. The algorithm will also have difficulty in
detecting horizontal gene transfer and other causes of
heterogeneity.
shows the plot of a program
using only the 3rd position bias information. These methods
depend on the accuracy of the codon frequency statistics of
already found genes. The algorithm will also have difficulty in
detecting horizontal gene transfer and other causes of
heterogeneity.



Next: Detection of Promoter Regions
Up: Detection of Coding Regions
Previous: ORFs as Markov chains
Peer Itsik
2000-12-25