Next: Parsimony
Up: Character Based Methods
Previous: Character Based Methods
The trivial solution to the phylogeny problem would be to enumerate over all possible trees and calculate the target function for each one. However, the number of non-isomorphic, labeled, binary, rooted trees containing n leaves, can be shown to be:
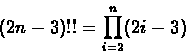 |
(1) |
(or (2n-5)!! for unrooted trees) which is of course super-exponential - for n=20, for instance, there are about 1021 such trees. This means that exhaustive enumeration is unfeasible even for a relatively small number of species.
The next sections will present several approaches towards defining a target function, and attempting to solve the problem for that target function.
Figure 8.1:
A most parsimonious 5-species phylogeny for a single DNA site. The bars mark the two possible edges along which a mutation might have occurred.
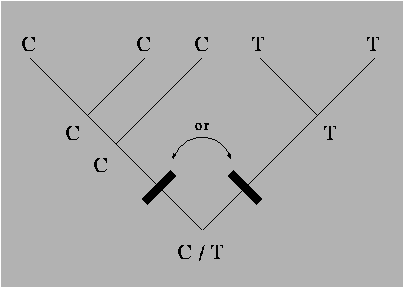 |
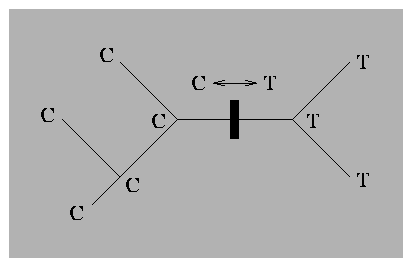
Figure 8.2:
The unrooted counterpart of the
phylogeny in figure 8.1. Notice that there is
now no ambiguity about the placement of the mutation.
 |



Next: Parsimony
Up: Character Based Methods
Previous: Character Based Methods
Peer Itsik
2001-01-01