- Fall 2012-2013 - Software 1 (TA)
- Spring 2012-2013 - Software 1 (TA)
- Fall 2013-2014 - Software 1 (TA)
- Spring 2013-2014 - Software 1 (TA)
- Fall 2014-2015 - Software 1 (TA)
- Spring 2014-2015 - Software 1 (TA)
- Fall 2015-2016 - Python Programming for Engineers (Lecturer)
- Spring 2015-2016 - Python Programming for Engineers (Lecturer)
- Spring 2015-2016 - Computer Programming - C for Chemists (Lecturer)
- Fall 2016-2017 - Python Programming for Engineers (Lecturer)
- Spring 2016-2017 - Python Programming for Engineers (Lecturer)
- Fall 2017-2018 - Python Programming for Engineers (Lecturer)
- Spring 2017-2018 - Python Programming for Engineers (Lecturer)
- Fall 2018-2019 - Python Programming for Engineers (Lecturer)
- Spring 2018-2019 - Python Programming for Engineers (Lecturer)
- Fall 2019-2020 - Python Programming for Digital Sciences for Hitech (Lecturer)
- Fall 2019-2020 - Python Programming for Communication (Lecturer)
- Fall 2020-2021 - Python Programming for Digital Sciences for Hitech (Lecturer)
- Fall 2020-2021 - Data Handling for Business Analytics (Lecturer)
1.
Induction in
myeloid leukemic cells of
genes that are expressed in different normal tissues.
Lotem J, Benjamin
H, Netanely D, Domany E, Sachs L.
Proc Natl Acad
Sci U S A. 2004 Nov 9;101(45):16022-7. Epub 2004 Oct 25.
2.
Network analysis
of protein structures
identifies functional residues.
Amitai G, Shemesh
A, Sitbon E, Shklar M, Netanely D, Venger I,
Pietrokovski S.
J Mol Biol. 2004
Dec 3;344(4):1135-46.
3.
Human cancers
overexpress genes that
are specific to a variety of normal human tissues.
Lotem J, Netanely
D, Domany E, Sachs L.
Proc Natl Acad
Sci U S A. 2005 Dec 20;102(51):18556-61. Epub 2005 Dec 8
4.
Age-dependent
spatial memory loss can
be partially restored by immune activation.
Ron-Harel N,
Segev Y, Lewitus GM, Cardon M, Ziv Y, Netanely D,
Jacob-Hirsch J, Amariglio N, Rechavi G, Domany E, Schwartz M.
Rejuvenation Res.
2008 Oct;11(5):903-13.
Sheyn D, Pelled G,
Netanely D, Domany E, Gazit D.
Tissue Eng Part
A. 2010 Nov;16(11):3403-12.
6.
Transcription
Factor/microRNA Axis Blocks Melanoma Invasion
Program by miR-211 Targeting NUAK1.
Bell RE, Khaled
M, Netanely D, Schubert S, Golan T, Buxbaum A,
Janas MM, Postolsky B, Goldberg MS, Shamir R, Levy C.
J Invest
Dermatol. 2013 Aug 9. doi: 10.1038/jid.2013.340.
7.
MicroRNAs of the
RPE are essential for RPE differentiation
and photoreceptor maturation.
Ohana R,
Weiman-Kelman B, Raviv S, Tamm E, Pasmanik-Chor M, Rinon A, Netanely
D, Shamir R, Salomon AS, Ashery-Padan R.
Development. 2015
Jul 15;142(14):2487-98. doi: 10.1242/dev.121533.
PMID: 26062936
Netanely D,
Avraham A, Ben-Baruch A, Evron E, Shamir R.
Breast Cancer
Res. BioMed Central; 2016;18:74.
9.
PROMO:
An interactive tool for analyzing clinically-labeled multi-omic cancer
datasets
Netanely D,
Stern N, Laufer I, Shamir R.
BMC
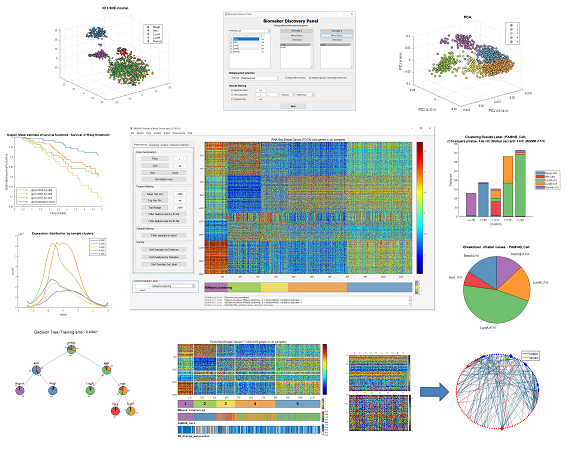
Bioinformatics. BioMed Central; 2019;20:732.
Santana-Magal N,
Farhat-Younis L, Gutwillig A, Gleiberman A,
Rasoulouniriana D, Tal L, Netanely D, Shamir R,
Blau R, Feinmesser M,
Zlotnik O, Gutman H, Linde IL, Reticker-Flynn NE, Rider P, Carmi Y.
Cancer Res. 2020
May 15;80(10):1942-1956. doi:
10.1158/0008-5472.CAN-19-2944.
PMID: 32127354
Netanely D,
Leibou S, Parikh R, Stern N, Vaknine H, Brenner R, Amar S, Factor RH,
Perluk T,
Frand J, Nizri E, Hershkovitz D, Zemser-Werner V, Levy C, Shamir R.
Oncogene.
2021 Feb 9. doi: 10.1038/s41388-021-01665-0. PMID: 33564068