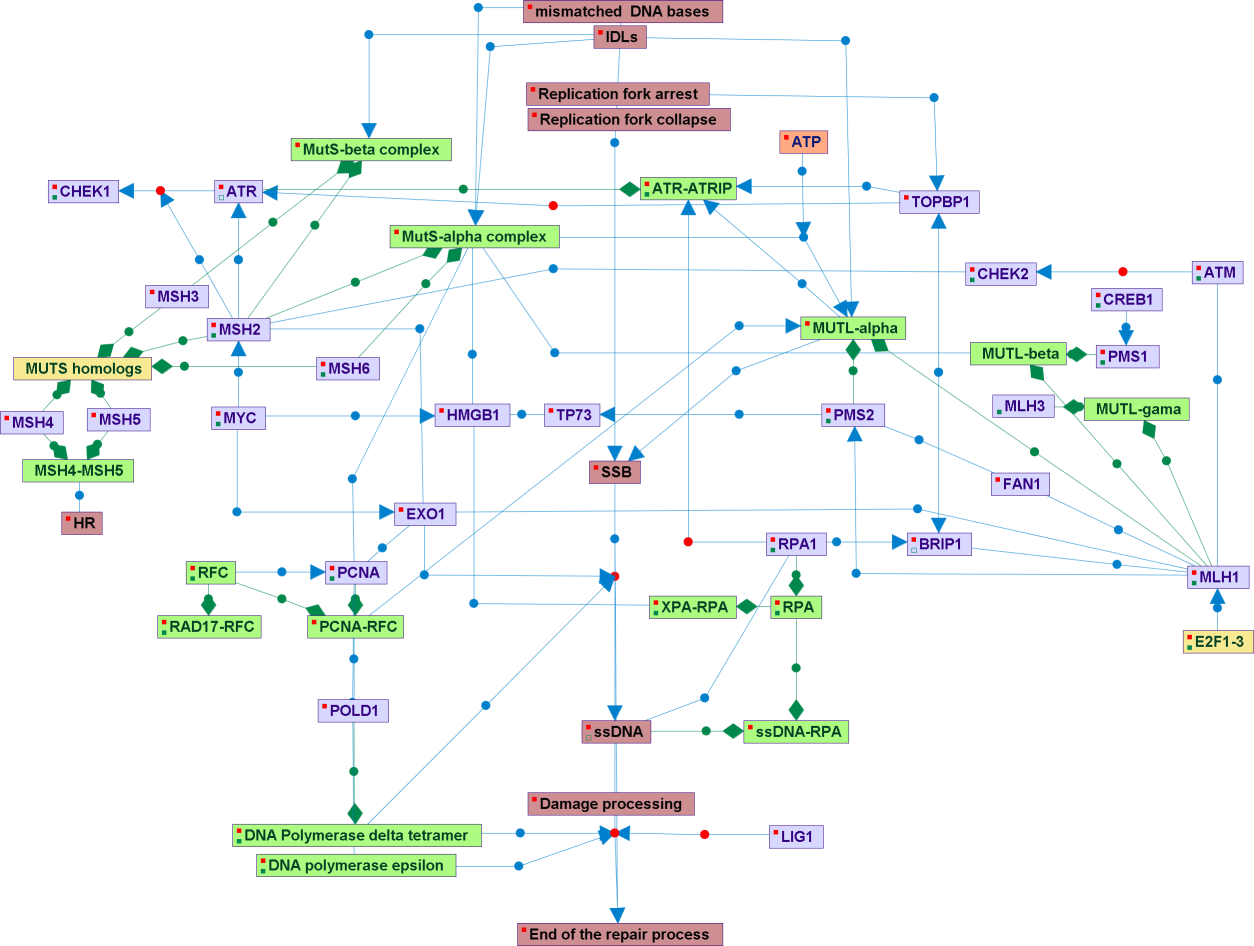
MMR is a system for recognizing and repairing erroneous insertions/deletions loops (IDLs), and mis-incorporation of bases that can arise during DNA replication and recombination, as well as repairing some forms of DNA damage. It primarily corrects single base-pair mismatches and small misalignments IDLs, which arise during replication. The MMR is strand-specific. As the newly synthesised (daughter) strand commonly includes errors, the MMR machinery distinguishes the newly synthesised strand from the template, excising the mismatch nucleotide from the nascent strand (generally together with a stretch of nucleotides neighboring the misaligned nucleotide). The number of the excised nucleotides can be up to a kilobase away from the misalignment - the ends of Okazaki fragments or the 3?-termini of the leading strands during replication. The termini of invading strands may fulfill this role during recombination. Mismatch repair (MMR) is an essential process to maintain genome integrity MMR increases the replication fidelity by up to three orders of magnitude. In humans mismatch recognition is mediated by one of two heterodimers, composed of the MutS homologs MSH2 and MSH6 (MutS-alpha) or MSH2 and MSH3 (MutS -beta). MutS-alpha which is more abundant, is involved in the repair of base/base mismatches and single nucleotide misalignments while MutS-beta recognizes larger IDLs. Mismatch bound MutS-alpha (or MutS-beta) recruits a second heterodimer referred to as MutL-alpha which is composed of the MutL homologs MLH1 and PMS2. In humans MutL-alpha possesses endonuclease activity localised to the PMS2 subunit enabling MutL-alpha to introduce random nicks at sites spanning the mismatch. Subsequent loading of EXO1 at the 5’ side of the mismatch leads to activation of its 5’–3’ exonuclease activity resulting in removal of the incorrect DNA-fragment. The remaining single-stranded gap is filled by polymerase delta and its cofactors proliferation cell nuclear antigene (PCNA) and replication factor C (RFC). The repair process is completed when the remaining nick is sealed by DNA ligase I.. The MMR can activate the cell cycle checkpoints through the ATM and ATR kjnases. The MutS-alpha complex and the MLH1 and HGMB1 proteins might be involved in additional repair processes beyond the MMR.