|
G1-S Phase of the Cell Cycle
Created by: Rani Elkon, Tel Aviv University
Created on: Aug, 2006
Last modified on: Jan, 2007
|
|
|
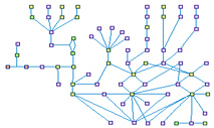
G1-S Phase of the Cell Cycle (spike00001)
The core engines that drive the progression through the eukaryotic cell cycle are complexes of cyclins and cyclin dependent
kinases. Cdk4-Cyclin D complex is implicated in committing a cell to the G1/S phase transition, Cdk2-Cyclin E in the
initiation of S phase and Cdk2-Cyclin A in its completion as well as in S phase exit. CDKN1A that is activated by various
kinds of damages to the cell, interact physically with the latter two aforementioned complexes, and inhibit the cell cycle
progression.
The E2F1-3 protein family members are the major transcription factors (TFs) that regulates the transcription of genes
involved in the cell progression from G1 to the S phase.
|
|
|
|
|
|
G2-M Phase of the Cell Cycle
Created by: Rani Elkon, Tel Aviv University
Created on: Aug, 2006
Modified by: Arnon Paz, Tel Aviv University
Last modified on: Jan, 2010
|
|
|
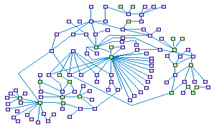
G2-M Phase of the Cell Cycle (spike00002)
The entry of a cell into mitosis is regulated by an elaborate network of kinases and phosphatases, that controls both the
timing of the division, and the complete reorganization of the cellular architecture. Cyclin/Cdk1(CDC2) is thought to be
the major kinase that initiates the onset of mitosis. The checkpoint kinases CHEK1 and CHEK2, when activated by ATM, mediated
the CycB-CDC2 complex activity by inhibiting its direct activator CDC25C. The transcription factor FoxM1, peaking in the late
G2 phase, directly enhance cyclin B transcription. Transcription factors and protein modifiers regulates the amount of the
components of the large multiprotein complexes involved in the mitosis progression (cohesin, centromer-kynetochore and the
mitotic spindle), their recruitment and the activity of these complexes.
|
|
|
|
|
|
DNA damage induced G1-S checkpoint (spike00003)
Created by: Arnon Paz, Tel Aviv University
Created on: Nov, 2010
|
|
|
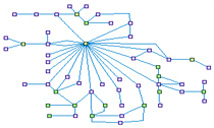
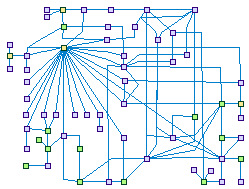
DNA damage induced G1-S checkpoint (spike00003)
DNA damage activates Ataxia telangiectasia mutated (ATM) and/or ATM and Rad3-related (ATR) signaling kinases, which trigger a dual wave of
checkpoint responses in the G1-S boundary: (i) A quick (but transient) CDK2/1 inhibition achieved by (ATM-activated)-CHEK2 or
(ATR-activated)-CHEK1 phosphorylation of the phosphatase CDC25A (the CDKs' activator), a phosphorylation that enhance the degradation of
CDC25A. (ii) A Slower, but sustained process depends on the transcription factor TP53. ATM/ATR phosphorylates (directly or indirectly)
both TP53 and MDM2 - the TP53 regulatory protein - which enhance TP53 stability and its DNA binding activity. TP53-mediated expression
of p21(CDKN1A) leads to CycA-CDK2, CycE-CDK2, and CycB-CDC2 (CDK1) complexes inhibition, halting the cell in G1.
|
|
|
|
|
|
Response to DSBs
Created by: Arnon Paz, Tel Aviv University
Created on: Jan, 2009
Last modified on: Mar, 2011
|
|
|
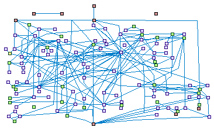
Response to DSB (spike00004)
DNA double-strand breaks (DSBs) arise in cells from and exogenous
and endogenous attacks on the DNA backbone. Ionizing radiation (IR)] is the main exogenous cause of DSB.
Proper repair of DSBs is critical and essential for maintaining genomic integrity and stability.
Eukaryotic organisms have evolved multi-potent and efficient mechanisms to repair DSBs that are primarily
divided into two types of pathways: nonhomologous end joining (NHEJ) and homologous recombination (HR).
|
|
|
|
|
|
Nucleotide excision repair
Created by: Arnon Paz, Tel Aviv University
Created on: Oct, 2009
Last modified on: Apr, 2011
|
|
|
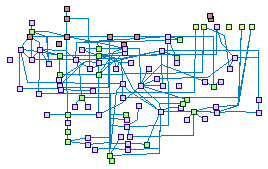
Nucleotide excision repair (spike00005)
Nucleotide excision repair (NER) is a particularly important
mechanism by which the cell can prevent unwanted mutations by removing the vast majority of UV-induced DNA
damage (mostly in the form of thymine dimers and 6-4-photoproducts). NER can be divided into two subpathways
(Global genomic NER and Transcription coupled NER) that differ only in their recognition of helix-distorting
DNA damage.
|
|
|
|
|
|
ATM Signaling Network
Created by: Arnon Paz, Tel Aviv University
Created on: Jan, 2009
Last modified on: Jan, 2012
|
|
|
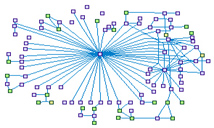
ATM Signaling Network (spike00006)
Ataxia telangiectasia mutated (ATM) is a serine/threonine-specific
protein kinase that is recruited and activated by DNA double-strand breaks (DSBs). It phosphorylates several key proteins that
initiate activation of the DNA damage checkpoint, leading to cell cycle arrest, DNA repair or apoptosis.
Several of these targets, including p53, CHK2, BRCA1, and H2AX are tumor suppressors.
|
|
|
|
|
|
Repair of Interstrand Crosslinks
Created by: Arnon Paz, Tel Aviv University
Created on: Jan, 2010
Last modified on: May, 2011
|
|
|
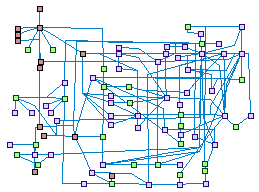
Repair of Interstrand Crosslinks (spike00007)
Multiple pathways of DNA repair participate in the repair of DNA
interstrand cross-links (ICLs). The major pathway is homologous recombination (HR) with involvement of the Fanconi
anemia/BRCA proteins. Components of the nucleotide excision repair (NER) and mismatch repair (MMR), has cardinal
roles in ICLs-sensing, signaling, and repair processes. Trans-lesion synthesis (TLS) by various error prone DNA
polymerases contributes also to the ICLs damage resolution.
|
|
|
|
|
|
Base excision and single strand break repair
Created by: Arnon Paz, Tel Aviv University
Created on: Mar, 2010
Last modified on: Jun, 2011
|
|
|
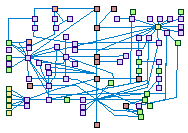
Base excision and single strand break repair (spike00008)
Base excision repair (BER) is primarily responsible for removing small, non-helix
distorting base lesions from the genome. These lesions are resulting from various chemical processes: oxygenations, deaminations,
alkylations, and spontaneous hydrolysis of the N-glycosidic bond of the bases. BER is initiated by DNA glycosylases, which
recognize and remove specific damaged or inappropriate bases, forming apurinic/apyrimidinic (AP) sites. These are then cleaved
by an AP endonuclease or AP DNA lyase. The resulting single-strand break can then be processed by either short-patch (SP-BER)
where a single nucleotide is replaced, or long-patch (LP-BER), where 2-10 new nucleotides are synthesized.by a DNA polymerase,
The final step of BER entails ligation of the remaining nick, by either LIG1 alone or LIG3–XRCC1 complex.
|
|
|
|
|
|
Mismatch repair (MMR)
Created by: Arnon Paz, Tel Aviv University
Created on: Mar, 2010
Last modified on: Jun, 2011
|
|
|
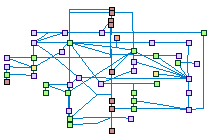
Mismatch repair (spike00009)
Mismatch repair (MMR) is a system for recognizing and repairing erroneous
insertions/deletions loops (IDLs), and mis-incorporation of bases that can arise during DNA replication and recombination,
as well as repairing some forms of DNA damage. It primarily corrects single base-pair mismatches and small misalignments IDLs,
which arise during replication.
|
|
|
|
|
|
DNA damage response and chromatin remodeling
Created by: Arnon Paz, Tel Aviv University
Created on: Oct, 2010
|
|
|
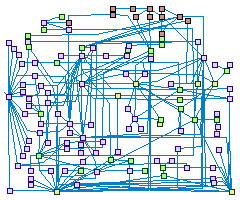
DNA damage response and chromatin remodeling (spike00010)
The packaging of eukaryotic chromosomal DNA into nucleosomes impedes the access of
the DNA repair machinery to damaged DNA. To deal with this obstacle, transient chromatin structural changes are integral to base-
and nucleotide-excision, homologous recombination, and non-homologous end joining DNA repair pathways, allowing better accessibility
of the DNA repair proteins to the damaged sites, and/or stabilize their recruitment. The chromatin remodeling mechanisms includes:
(1) posttranslational modifications (phosphorylations. acetylations, ubiquitination etc.) of histones and non-histones chromatin
associated proteins (2) histone-variants replacement, and nucleosomes repositioning or eviction. ATP-dependent multi subunits
chromatin remodeling complexes (INO80, SRCAP/SWR-C, SWI/SNF etc.) implement the latter mechanisms.
|
|
|
|
|
|
Apoptosis
Created by: Yaara Ber, Weizmann Institute of Science
Created on: Jan, 2009
Last modified on: Apr, 2010
|
|
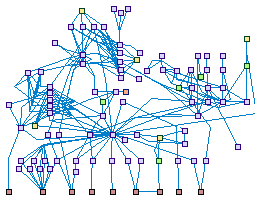
|
Apoptosis (spike00011)
Apoptosis is a genetically controlled program by which the cell
commits suicide in response to external or intenal stimuli, thereby eliminating excess, damaged and otherwise
unwanted cells. It is mediated by the caspase family of proteases, which cleaves multiple cellular substrates,
leading to the morphologic changes that characterize the apoptotic cell. These include fragmentation of the
cytoplasm and nucleus, chromatin condensation and fragmentation, cytoskeletal collapse, membrane blebbing,
and finally, disintegration of the cell into apoptotic bodies, which are engulfed by adjacent cells.
|
|
|
|
|
|
Caspases Cascade
Created by: Shani Bialik Brown, Weizmann Institute of Science
Created on: Oct, 2009
Last modified on: Oct, 2009
|
|
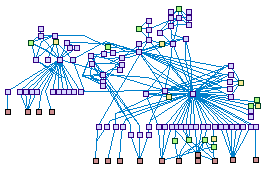
|
Caspases Cascade (spike00012)
Caspases are a family of cysteine proteases that serve as the
executioners of apoptosis. Cleavage of caspase substrates leads to the orderly dismantling of the cell which
characterizes apoptotic cell death.
|
|
|
|
|
|
DAPk family
Created by: Shani Bialik Brown, Weizmann Institute of Science
Created on: Oct, 2009
Last modified on: Oct, 2009
|
|
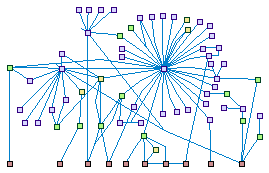
|
DAPk family (spike00013)
The tumor suppressor Death
Associated Protein Kinase (DAPk, DAPK1) is a Ser/Thr kinase with multiple substrates that regulates several forms
of cell death, including apoptosis, autophagic cell death, and programmed necrosis. It belongs to a family of
closely related kinases, including DRP-1 (DAPK2) and ZIPk/Dlk (DAPK3), which share similar functions.
|
|
|
|
|
|
Apoptosis Anti-Apoptosis Network
Created by: Arnon Paz, Tel Aviv University
Created on: Jan, 2009
Last modified on: Jan, 2012
|
|
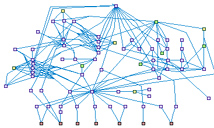
|
Apoptosis Anti-Apoptosis Network (spike00014)
Apoptosis is a mode of cell death that plays an important role
in both pathological and physiological processes. When cells exposed to a trigger (apoptotic stimulus),
the apoptotic cascade starts. There are several proteins involved in cell survival, proliferation, and
protection from apoptosis. When a cell is attacked by an apoptotic stimulus, the cell responds first by
activating anti-apoptotic mechanisms, which may or may not be followed by apoptosis. Whether or not a cell
undergoes proliferation, the survival, or apoptosis, appears to involve a balance between the two mechanisms.
|
|
|
|
|
|
Autophagy
Created by: Yaara Ber, Weizmann Institute of Science
Created on: Jul, 2009
Last modified on: Aug, 2011 By Shani Bialik
|
|
|
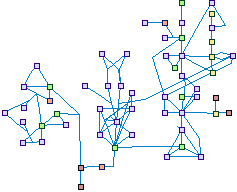
Autophagy (spike00015)
Autophagy, or “self eating”, is the process by which de novo-formed
double-membrane enclosed vesicles, known as autophagosomes, engulf cytosolic components and organelles, and
ultimately fuse with the lysosome, enabling degradation of its internal contents. Autophagy serves as a means of
recycling critical cellular building blocks, especially in times of deprivation and stress, and removal of damaged
organelles, and misfolded and aggregated proteins. The various stages of autophagosome formation are mediated by
the Atg genes, and induction of autophagy is modulated by several signaling pathways and regulators.
|
|
|
|
|
|
p53 Signaling Network
Created by: Rani Elkon, Tel Aviv University
Created on: Aug, 2006
Modified by: Arnon Paz, Tel Aviv University
Last modified on: Jan, 2012
|
|
|
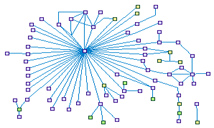
p53 Signaling Network (spike00016)
This gene encodes tumor protein p53, which responds to diverse
cellular stresses to regulate target genes that induce cell cycle arrest, apoptosis, senescence, DNA repair,
or changes in metabolism.
|
|
|
|
|
|
NFkB Signaling Network
Created by: Rani Elkon, Tel Aviv University
Created on: Aug, 2006
Modified by: Arnon Paz, Tel Aviv University
Last modified on: Jun, 2011
|
|
|
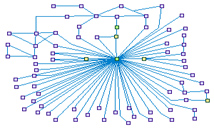
NFkB Signaling Network (spike00017)
NFkB (nuclear factor kappa-light-chain-enhancer of activated B cells)
is a protein complex that controls the transcription of DNA. NFkB is involved in cellular responses to stimuli
such as stress, cytokines, free radicals, ultraviolet irradiation, oxidized LDL, and bacterial or viral antigens.
NFkB plays a key role in regulating the immune response to infection, and also has an important anti-apoptotic
role.
|
|
|
|
|
|
Hearing related SIX1 Interaction
Created by: Zippi Brownstein, Tel Aviv University
Created on: Jun, 2009
Last modified on: Jun, 2009
|
|
|
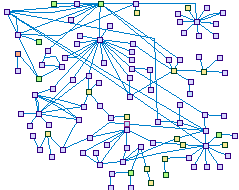
Hearing related SIX1 Interaction (spike00020)
The PAX, EYA, SIX and DACH gene families are important in otic
development. The SIX1 network shows the connections between them and further interactions diverging from SPIKE,
which were confirmed to be expressed in the ear.
|
|
|
|
|
|
MYO7A Interactions In The Ear
Created by: Zippi Brownstein, Tel Aviv University
Created on: Jun, 2009
Last modified on: Jun, 2009
|
|
|
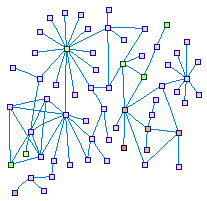
MYO7A Interactions In The Ear (spike00021)
Defects in myosin VIIA, as well as in other interactors of the
myosin VIIA network, are involved in Usher syndrome and in non-syndromic hearing loss. The map shows the
connections between the genes reported in the hearing system and other interactions diverged from the genes
inserted.
|
|
|
|
|
|
NOTCH1 Signaling In The Ear
Created by: Zippi Brownstein, Tel Aviv University
Created on: Jun, 2009
Last modified on: Jun, 2009
|
|
|
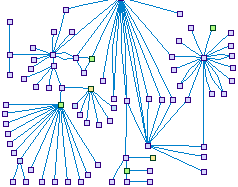
NOTCH1 Signaling In The Ear (spike00022)
In the ear, the Notch signaling pathway is involved in cell fate
decisions and in controlling the pattern of sensory hair cells and supporting cells through lateral
inhibition.
|
|
|
|
|
|
MYO3A Interacting Proteins in the Auditory System
Created by: Zippi Brownstein, Tel Aviv University
Created on: Aug, 2010
|
|
|
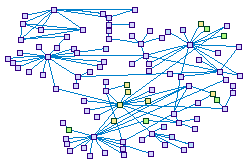
MYO3A Interacting Proteins in the Auditory System (spike00023)
Data from mass spectrometry analysis done in the Avraham's lab was combined into
HearSpike. Most of the inserted interacting proteins are new in the inner ear while some of them, like espin, fus and BCL2 are known
to interact in other pathways in the auditory system.
|
|
|
|
|
|
Apoptosis In The Ear
Created by: Zippi Brownstein, Tel Aviv University
Created on: Aug, 2010
|
|
|
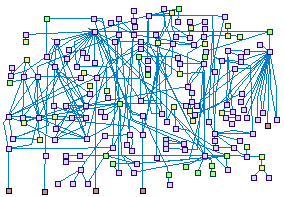
Apoptosis In The Ear (spike00024)
Cell death in inner ear tissues is an important mechanism leading to hearing
impairment. Members of several protein families such as SOD, Bcl-2, Casp and Calp contribute substantially to cell death in
the inner ear. All these proteins and many more act together in an intricate network, as is shown in the HEarSpike map.
|
|
|
|
|
|
Hearing and Vision Proteins
Created by: Zippi Brownstein, Tel Aviv University
Created on: Aug, 2010
|
|

|
Hearing and Vision Proteins (spike00025)
The genes related to Usher syndrome may shed light on proteins that play important
roles in normal hearing, as well as in vision. The protiens encoded by genes related to Usher syndrome, e.g.,
MY07A, USH1C, CDH23, PCDH15, SANS, as well as other proteins expressed both in the ear and in the eye, and the interactions
between them, are shown on the 'Hearing and Vision Protein' map.
|
|
|
|
|
|
The GJB2 Interactions in the Ear
Created by: Zippi Brownstein, Tel Aviv University
Created on: Feb, 2011
|
|
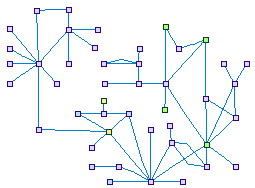
|
The GJB2 Interactions in the Ear (spike00026)
GJB2, encoding connexin 26 protein, is involved in up to 50% of non-syndromic
hearing loss in many world populations. Interactions with other connexins are known, and recent studies report interactions with
the supporting cell proteins in the organ of Corti known as the epithelial support complex (ESC).
|
|
|
|
|
|
WNT signaling
Created by: Arnon Paz, Tel Aviv University
Created on: Nov, 2011
|
|
|
WNT signaling (spike00027)
The Wnt pathway plays a critical role in the development of multicellular organisms.
Wnt family of proteins in human comprises a group of 19 secreted lipid-modified glycoproteins that play a crucial role in the
regulation of embryogenesis, development, cell proliferation, differentiation, polarity, migration, and invasion. Abnormal Wnt
signaling has been associated with many human diseases, including bone density defects, limb malformations, and cancer, There
are at least three different Wnt pathways: the canonical pathway, the planar cell polarity (PCP) pathway and the Wnt/Ca2+ pathway.
A critical component of several Wnt-signaling branches is the cytoplasmic protein Dishevelled (DVL).
|
|
|
|
|
|
RAS signaling and tumorigenesis
Created by: Arnon Paz, Tel Aviv University
Created on: Jan, 2012
|
|
|
RAS signaling and tumorigenesis (spike00028)
The RAS GTPases (HRAS, KRAS,and NRAS) signaling pathway plays a key role in the
regulation of cell cycle through the activation of numerous downstream pathways including the RAF-MEK-ERK, PI3K/AKT/mTOR, and
RAL pathways. RAS is the central hub in the growth factors signaling cascades leading to proliferation, and some of the RAS
effectors also mediate anti-apoptotic processes Aberrant regulation of RAS may lead to oncogenic transformation. Later,
changes in the RHO GTPases signaling within the tumor cells may promote their invasion, migration, and metastasis.
|
|
|
|
|
|
|
Contact
Questions? Please e-mail us at spike@post.tau.ac.il.
|
|
|