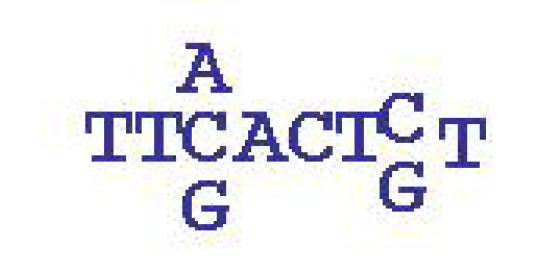Tel Aviv University School
of Computer Science

Fall 2013-14
Workshop in Computer Science
0368-3500-04
http://www.cs.tau.ac.il/~rshamir/workshop/13/
Workshop instructor: Prof. Ron Shamir
Lab Instructor: Yaron Orenstein (yaronore AT post.tau.ac.il)
Workshop: Kaploon 205, Tuesdays 15-17 ; Lab: Tuesdays 17-20
Downloads:
Test2 files ,
Test1 files ,
Training files ,
Motif logo
Workshop Topic: The workshop will deal with
design, analysis and development of efficient algorithms for finding
sequence motifs in high-throughput SELEX (HT-SELEX) data.
The motivation comes from identifying regulatory motifs in DNA, an important topic that
has been under intensive research for over ten years.
HT-SELEX is a new type of experimental data that has become available on a large scale
only very recently, and it open new opportunities to develop accurate motifs based
on very large data sets of sequences.
As part of the project,
application of the software developed on real biological data will be performed.
The high-throughput SELEX process is described in the following paper:
Jolma et al.
Multiplexed massively parallel SELEX for characterization of human
transcription factor binding specificities.
Genome Research, 2010.
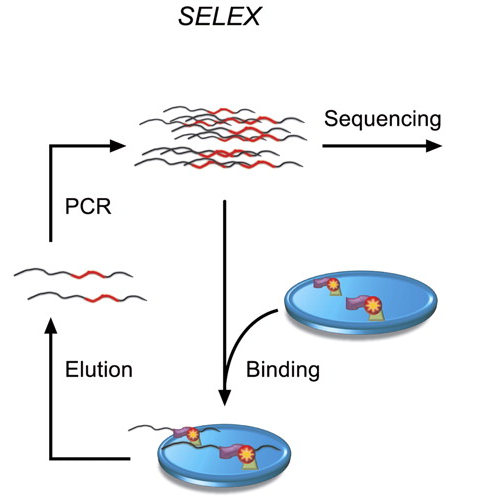
The output of the process looks like this:
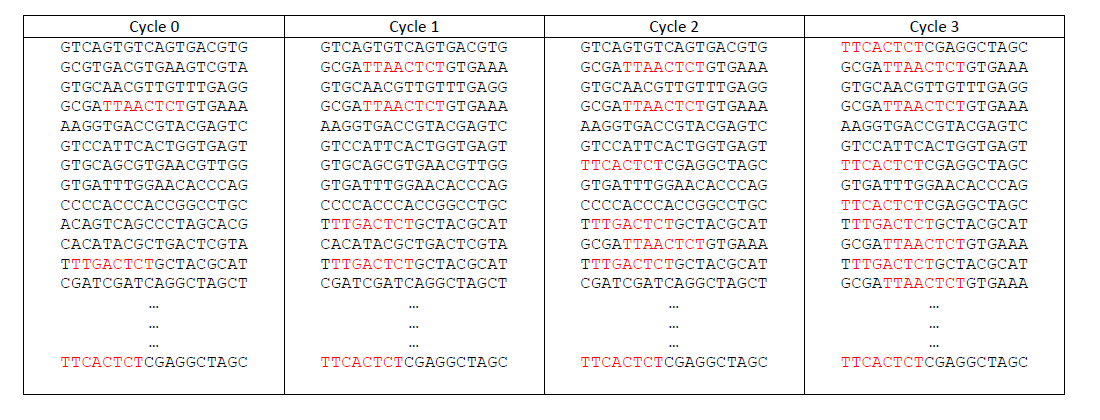 This is a simplified example of HT-SELEX dataset. It
consists of 4 files of ~100000 sequences, each of length 18 in the four-letter DNA.
The first cycle consists of random sequences.
This is a simplified example of HT-SELEX dataset. It
consists of 4 files of ~100000 sequences, each of length 18 in the four-letter DNA.
The first cycle consists of random sequences.
The sequences that contain the motifs are enriched from cycle to cycle.
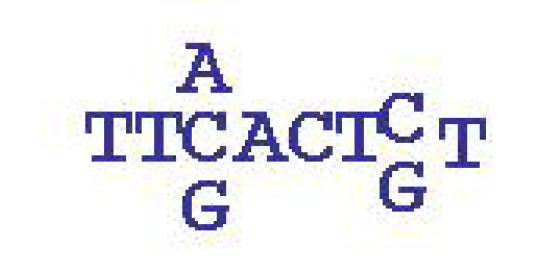
A simplified example of a motif: it is a sequence of length 6-12, which is typically
degenerate: in some positions alternative letters may occur. The motif shown is of
length 8 where the 3rd and 7th positions are degenerate. Occurrences of this motif is marked in
red in the HT-SELEX sequences above.
In more general motif models, each of the four letters
has a specified probability in each position.
For a survery of motif finding algorithms:
Das and Dai.
A survery of DNA motif finding algorithms.
BMC Bioinformatics, 2007.
Prerequisites:
The workshop is open to all 3rd year students in computer science. No biological
background is assumed. In case the workshop is oversubscribed, there will be
preference to students in the bioinformatics track. Knowledge of Java is required.
Format:
The work will be done by pairs of students or individually. We shall have 2-3
introductory meetings in the beginning of the semester to provide the necessary
background. Then groups will be formed and each group will start the design phase of
its project. After individual meetings with the groups and confirmation of the design,
the implementation will start. Towards the end of the semester, joint meetings of all
participants will take place, in which each group will present its project. After the
completion of the project, each group will meet with the instructors to demonstrate the
software and evaluate its performance, in addition to submitting the results of the
algorithm on the test data.
Consultation meetings of single groups with the instructors will be carried out
throughout the semester as needed and individual meeting times will be set.
The assigned time slots of the lab (MAABADA) are a formality, and will not be used.
Students will be given training datasets with given solutions, for training and
practice, and test datasets. The same datasets will be given to all groups. The
performance of all algorithms will be measured on the test datasets. In addition, in
the final meeting with the instructors, an additional dataset will be given for online
query testing.
Software: The algorithms will be implemented
in JAVA and tested on Linux.
Grading:
-
15% for the design
-
25% for the implementation (10% for modularity, clarity, documentation, f(r,k)*15% for
efficiency)
-
f(r,k)*50% for the accuracy of the test results
- f(r,k)*15% for test1
- f(r,k)*20% for test2
- f(r,k)*15% for test3
where r= group's rank in test out of k groups (top rank r=k) and f(r,k)=0.5+0.5*r/k
-
20% for the final report and presentation
-
5% bonus to the group with the most accurate results
-
5% bonus for the group with the fastest implementation
Schedule:
-
19/11 First progres report (meetings)
-
10/12 Test1 (submission)
-
24/12 Design document (submission)
-
14/1 Test2 + executable (submission)
-
18/2 Final presentation (meeting)
Slides: Introduction (15/10)
Slides: background and project plan
(22/10)