Tel Aviv University School of Computer Science

Tel Aviv University School of Computer Science

Downloads: Training files , Fixed training files , Test files , Math functions , Motif logo , ComparePwms.java First assignment: Research report, due 23/11/10. Workshop Topic: The workshop will deal with design, analysis and development of efficient algorithms for finding sequence motifs in Protein Binding Microarray (PBM) data. The motivation comes from identifying regulatory motifs in DNA, an important topic that has been under intensive research for over ten years. As part of the project, application of the software developed on real biological data will be performed.
|
Sequence |
Signal |
|
CATGTAAGAGTTGACTCTGGTCTGTTCTAAT |
28926 |
|
TTGCTCATCAGAGTCGCGTAACAGGCTTTC |
1457 |
|
TCCAGTTTAGGTGGCGCCCGGAACCCTTAA |
12972 |
|
.. |
.. |
|
CATGTAGCCCTTAACTGTGACTAAAGCCCC |
33755 |
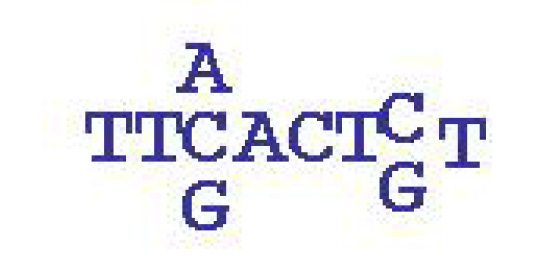
A simplified example of a motif: it is a sequence of length 6-12, which is typically
degenerate: in some positions alternative letters may occur. The motif shown is of
length 8 where the 3rd and 7th positions are degenerate. Occurrences of this motif is marked in
red in two of the BPM sequences above.
In more general motif models, each of the four letters
has a specified probability in each position.
Slides: background and project plan