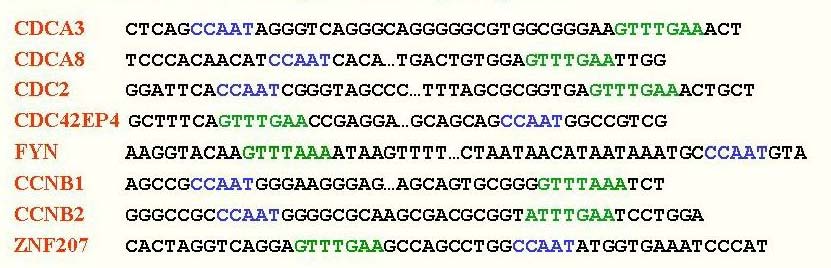
In this example, two sequence patterns (the blue and the green motifs) are marked in
eight different sequences.
For the biologically knowledgeable, each line shows a fragment of a promoter of a human
G2+M cell cycle gene (The gene names are lister on the left). Each such sequence is >1000 letters long. The blue motif (CCAAT) is the NF-Y recognition site, and the green
motif [G/A]TTT[G/A]AA is the CHR recognition site. It is clear from the picture that
these motifs tend to co-occur in close proximity in the same targe genes,
and indeed the transcription factors that they represent together regulate the expression of G2+M
cell cycle genes.