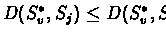Next: Common Multiple Alignments Methods
Up: Multiple Alignment to a
Previous: Multiple Alignment to a
Lifted Alignment tree - a Heuristic for
Phylogenetic Alignment
Definition 5.13
A phylogenetic alignment is called
lifted alignment if for
every internal node
v, the string assigned to
v is also
assigned to one of
v's children (See figure
5.2). Trivially, in such a case, all
internal nodes are assigned labels from the set of leaf strings.
Let T* be the optimal alignment for tree T. We will construct
a lifted alignment
TL = Lift(T*), which is based on T*,
with only a limited damage to the alignment distance. Note that
this construction is only conceptual since we usually do not know
T*.
For each node v let its T* label be S*v. We shall assign
every v a label SLv. Initially, only the leaves are labeled,
and by definition
SLv = S*v for each leaf v. The labeling
process successively traverses the internal nodes in any order,
provided that a node is not visited before any of its children.
Upon visiting a node, it is lifted i.e. labeled by one of
the labels of its children. Thus, the resulting phylogenetic
alignment is lifted. (see figure
5.3)
Procedure Lift(T : Tree)
begin
while there
exists an unlifted node v, all of whose children have been
lifted, do :
Find a child j whose label Sj is the
closest to S*v. Namely
For every child i of
v:
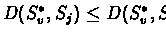
Label S*v with Sj
end while
end
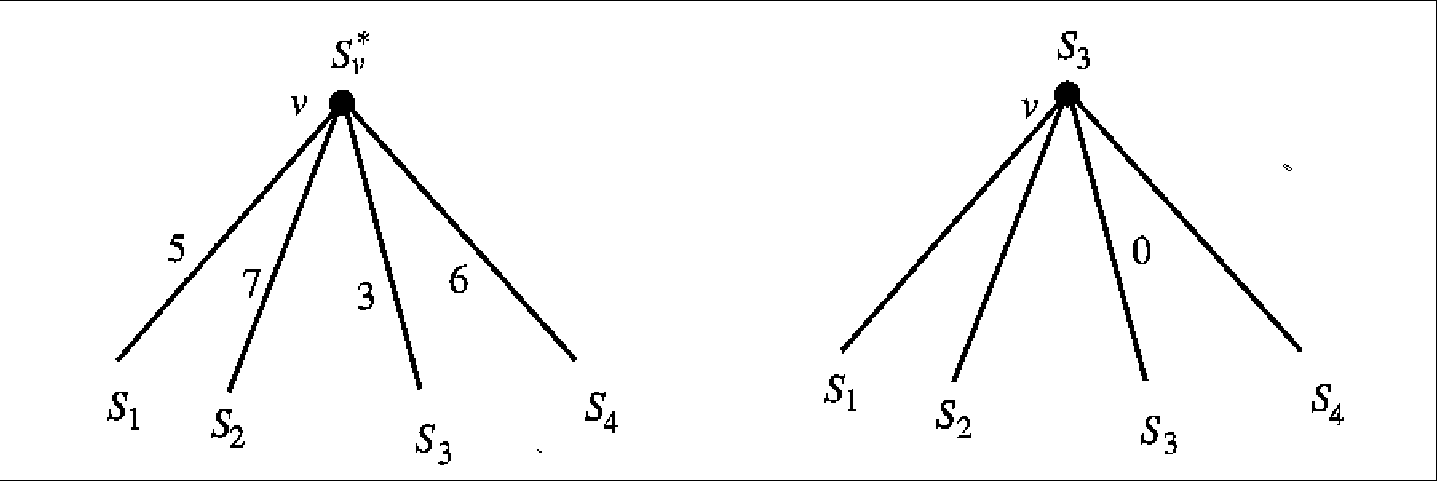
Figure 5.3:
The lifting construction at node
v. The numbers on the edges are the distances from S*v
to the lifted strings labeling its children. On the left is the
tree before lifting, and on the right the result of the lift.
After the lift one edge will have a distance of
0.
|
|
Theorem 5.7 (Jiang, Wang and Lawler, 1996 [
5])
The
distance of the phylogenetic alignment
TL =
Lift(
T*) is at
most twice the distance of the optimal phylogenetic alignment
T*.
Proof:Let e = (v,w) be an edge in T. Suppose
that in TL, Sj is the label of v and Si is the label of
w. If i=j then
D(Sj, Si) = 0. Otherwise:
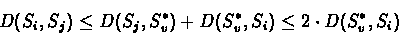 |
(5.8) |
The first inequality is due to the triangle inequality, and the
second follows from the labeling algorithm. For an edge e =
(v,w), with
Sw = Si ,Let Pe be the path in T from
v to the leaf labeled Si. Due to the triangle inequality
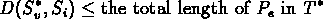 |
(5.9) |
We say that the edge e = (v,w) is blue in TL if
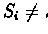 .
The distance of a lifted alignment TL is equal to
the sum of edge distances on all the blue edges in the tree.
.
The distance of a lifted alignment TL is equal to
the sum of edge distances on all the blue edges in the tree.
For a blue edge e = (v,w), observe that the definition
of lifted alignment implies that along the path Pe every node
except v is labeled Si, and no node outside Pe is labeled
Si. Hence, if
e' = (v', w') is any other blue edge, then
Pe and Pe' have no edges in common. This defines a mapping
from every blue edge e in TL to a path Pe in T* such
that:
- The distance in TL of the edge e is at most twice the
total distance in T* of the edges on Pe. (Follows from
equations 5.8 and 5.9.
- No edge in T* is mapped to by more than one edge
in TL.
Therefore the total distance of TL = the total distance on blue
edges 
 the sum of all total distances of Pe
paths in T*
the sum of all total distances of Pe
paths in T* 
 the total distance of T*. (see
figure 5.4)
the total distance of T*. (see
figure 5.4)
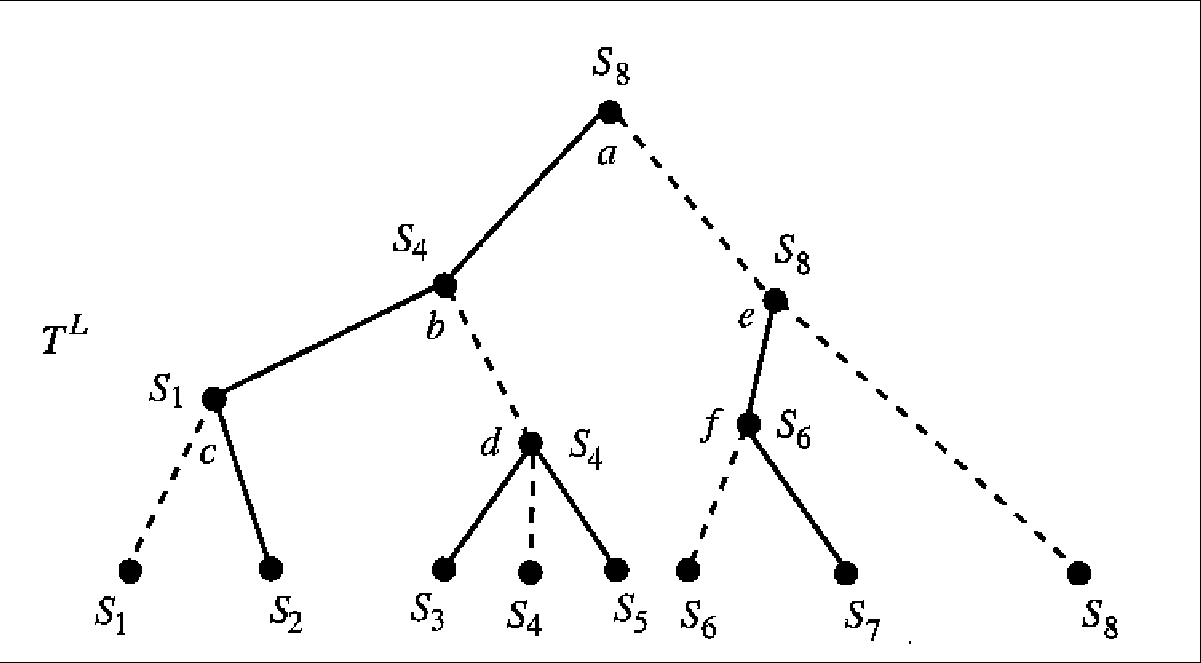
Figure 5.4:
The lifted tree T*L. The dashed
edges show the paths along which a leaf string has been lifted to
some internal node. Solid edges are blue edges in T*L, while
each of these dashed edges has distance 0. The path P(a,b)
for example, is the path b,d, S4 along which the string
labeling b was lifted. Edge (a,b) has distance in T*L at
most twice the distance of path P(a,b) in
T*.
|
|
We now describe how to find the optimal lifted alignment using a
dynamic programming algorithm as listed below. But first we
define:
Definition 5.14
Let
Tv be the subtree of
T rooted at node
v and
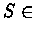
.
Let
d(
v,
S) denote the distance of the best lifted alignment of
Tv under the requirement that string
S is assigned to node
v.
The algorithm will compute d(v,S) for any 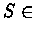 working it's way from the leaves up using the following recursion:
working it's way from the leaves up using the following recursion:
- If v is an internal node with all its children being
leaves, then
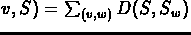 where Sw is the label of w.
where Sw is the label of w.
- Else,
![$d(v,S) = \sum_{(v,v')} min_{S' \in S} [D(S,S') + d(v',S')]$](img70.gif) where v' is a child of v and S' is a label of one of the leaves of Tv'.
where v' is a child of v and S' is a label of one of the leaves of Tv'.
Time analysis: We perform a preprocessing
stage, in which we compute all the
 pairwise
distances between the k input strings. This takes O(N2) time,
where N is the total length of all the strings. The work at any
internal node is O(k2), and the overall work of the algorithm
is
O(N2 + k3)
pairwise
distances between the k input strings. This takes O(N2) time,
where N is the total length of all the strings. The work at any
internal node is O(k2), and the overall work of the algorithm
is
O(N2 + k3)



Next: Common Multiple Alignments Methods
Up: Multiple Alignment to a
Previous: Multiple Alignment to a
Itshack Pe`er
1999-03-16