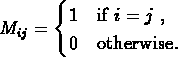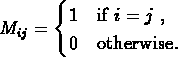Next: PAM units and PAM
Up: No Title
Previous: PSI BLAST - Position
Amino Acids Substitution Matrices
When we search for the best alignment between two protein sequences, the scoring (or substitution) matrix we use can largely
affect the results we get. Ideally, the scores should reflect the biological phenomena that the
alignment seeks to expose. For example - in the case of sequence divergence due to evolutionary mutations, the
values in the scoring matrix should ideally be derived from empirical observation on ancestral sequences and
their present-day descendants.
Two examples of simple substitution matrices often used that do not employ the biological phenomena are:
- 1.
- The unit matrix:
- 2.
- The genetic code matrix. Mij equals the number of minimal base substitution needed to convert a codon
of amino acid i to a codon of amino acid j. We disregard here the importance of chemical properties of the amino acids,
that evidently influence the chances for their substitution, like their hydrophobicity, charge or size.
Itshack Pe`er
1999-01-10