


Next: Clustering Using BioClust
Up: The BioCluct Clustering Algorithm
Previous: Algorithm Correctness
Based on ideas of theoretical algorithm given in previous sections, a simple and practical heuristic was implemented. All the tests described in subsequent sections were performed using this practical implementation of the theoretic algorithm.
Let C be a cluster. Let Si,j be a similarity matrix and let 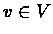 be a gene. We define the affinity of v to cluster C by
be a gene. We define the affinity of v to cluster C by
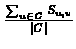 .
Given affinity threshold
.
Given affinity threshold 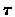 we will say that v is a close gene to cluster C if its affinity to C is above
we will say that v is a close gene to cluster C if its affinity to C is above 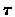 and we will say that v is a weak gene in C if its affinity to C is below
and we will say that v is a weak gene in C if its affinity to C is below 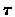 .
.
Following are the steps of the practical implementation:
- Construct one cluster at a time and denote it by CC;
- At each step either:
- Add a close gene to CC;
- Remove a weak gene from CC
- Close CC when no move is possible;
- Repeat until all genes are clustered;
The main differences between the practical implementation and the theoretical algorithm are:
- In the theoretical algorithm several partitions are formed and then the ``best'' partition is chosen; in the practical implementation one partition is formed by building one cluster at a time;
- The theoretical algorithm considers the similarity graph, while the practical implementation processes the similarity matrix (the similarity value between any two genes can assume any real value);
- In the theoretical algorithm, the clusters in a partition are extended by adding new elements to them; the practical implementation allows also to remove a weak element from a cluster;
Although nothing can be proved about the running time and performance of the practical implementation, the test results described in the next sections show that it performs remarkably well both on simulated data and on real biological data.



Next: Clustering Using BioClust
Up: The BioCluct Clustering Algorithm
Previous: Algorithm Correctness
Peer Itsik
2001-02-01
![]() be a gene. We define the affinity of v to cluster C by
be a gene. We define the affinity of v to cluster C by
![]() .
Given affinity threshold
.
Given affinity threshold ![]() we will say that v is a close gene to cluster C if its affinity to C is above
we will say that v is a close gene to cluster C if its affinity to C is above ![]() and we will say that v is a weak gene in C if its affinity to C is below
and we will say that v is a weak gene in C if its affinity to C is below ![]() .
.