


Next: Probabilistic Assumptions
Up: The CLICK Algorithm
Previous: The CLICK Algorithm
CLICK (CLuster Identification via Connectivity Kernels) is a newer algorithm
for clustering [20]. The input for CLICK
is the gene expression matrix. Each
row of this matrix is an ``expression fingerprint'' for a single gene. The
columns are specific conditions under which gene expression is measured (e.g.
different points in time). A more formal definition is as follows:
Let
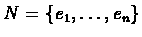 be a set of elements. Let M be an input real-valued matrix of order
be a set of elements. Let M be an input real-valued matrix of order 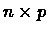 ,
where Mij is the j-th attribute of ei.
The i-th row-vector in M is the fingerprint of ej. For a set of elements
,
where Mij is the j-th attribute of ei.
The i-th row-vector in M is the fingerprint of ej. For a set of elements
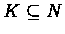 ,
we define the fingerprint of K to be the mean vector
of the fingerprints of the members of K. One seeks to partition N into
clusters (subsets). In such a partition, elements in the same cluster are
called mates.
The CLICK algorithm attempts to find a partition of N into clusters,
so that two criteria are satisfied: Homogeneity -
mates are highly similar to each other;
and separation - non-mates have low similarity
to each other.
,
we define the fingerprint of K to be the mean vector
of the fingerprints of the members of K. One seeks to partition N into
clusters (subsets). In such a partition, elements in the same cluster are
called mates.
The CLICK algorithm attempts to find a partition of N into clusters,
so that two criteria are satisfied: Homogeneity -
mates are highly similar to each other;
and separation - non-mates have low similarity
to each other.
Peer Itsik
2001-01-31