Next: The HCS Algorithm
Up: cDNA Clustering
Previous: Motivation
Recall that a gene is transcribed into mRNA, which is then translated into a
protein. In order to check what genes are expressed in a given tissue, we use
cDNA - a reverse-transcript of the mRNA, which is more stable. There exists
methods which enable us to extract cDNA in large quantities from the tissue,
and we can see, at a given moment, which cDNA molecules exist in the tissue
(details are omitted).
In fact, we sample cDNA molecules from the tissue. The more a gene is
expressed, the more samples of its matching cDNA we will find. The sample we
have obtained contains about 100,000 cDNA fragments, each of them between 500
and 2,500 base-pairs long, the average being around 1,200.
Reverse transcription of mRNA uses a poly-T primer that hybridizes to the
poly-A tail of the mRNA. All cDNA fragments we have from the gene will
thus have a common start. Since reverse transcription mya stop abruptly,
the length of such fragments may vary.
We can now formulate the problem we face:
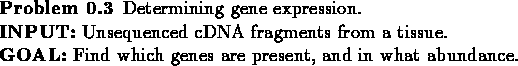
The simple solution is to sequence all the cDNA fragments we have extracted
from the tissue. This is both wasteful and slow. We have extracted a very
large quantity of cDNA, and many fragments come from the same genes.
Sequencing all of them will mean sequencing the same genes over and over
again.



Next: The HCS Algorithm
Up: cDNA Clustering
Previous: Motivation
Peer Itsik
2001-01-31