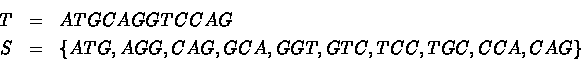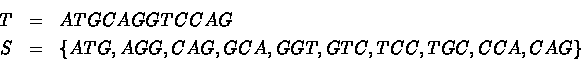Next: The naive approach
Up: DNA Chips/Microarrays
Previous: Manufacturing Oligonucleotide Arrays
Standard oligo chips can, at least theoretically, be used for sequencing. Let
us prepare an oligo chip that contains all possible sequences of length k.
These sequences are called k-mers. Practical values of k are 8-10.
If we expose this chip to a solution containing some target DNA, the results
will show which k-mers occur in the target sequence. This gives rise to the
definition of the k-spectrum of a sequence T as the multi-set
of all its substrings of length k. We would now like to reconstruct this
sequence.

It should be noted that we assume that if a k-mer appears several times in
the target DNA, the hybridization experiment will report its multiplicity
(this is why we require the input to be a multi-set and not simply a set). To
date, this requirement is impractical.
For instance, for k=3:
Peer Itsik
2001-01-31