Next: cDNA Microarrays
Up: DNA Chips/Microarrays
Previous: Oligo-Fingerprinting
The basic idea in these chips is similar. The difference is that these chips
contain the oligos, and not the targets. The length of the oligos used
depends on the application, but they are usually no longer than 25 bases.
Since oligos are usually shorter, the density is much higher in these chips.
For instance, a chip that is 1cm by 1cm can easily contain 100,000 oligos.
The chips are used in a similar manner. They are exposed to a solution
containing many copies of the target DNA. Hybridization occurs between oligos
and matching DNA and then the chip can be heated in order to repeat the
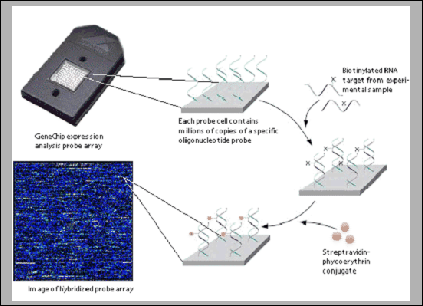
experiment. Figure 11.1 shows a schematic description of
such an experiment.
While the basic idea is the same, the chips are obviously intended for
different experiments. This kind of chips is most useful in situations
where we have relatively few targets, but a large number of oligos we wish to
test. Another advantage is that we may want to hybridize targets against a
``standard'' oligo library. The oligo chips can thus be manufactured in large
quantities, and their cost decreases.
Figure 11.1:
A typical experiment with an oligonucleotide chip.
Labeled RNA molecules are applied to the probes on the chip,
creating a fluorescent spot where hybridization has occurred
|
|



Next: cDNA Microarrays
Up: DNA Chips/Microarrays
Previous: Oligo-Fingerprinting
Peer Itsik
2001-01-31