


Next: Map Quality
Up: Constructing Physical Maps from
Previous: Clone Pair Overlap Score
- 1.
- Initialize a collection of contigs, each of which is a set of clones. Initially, the collection includes for each clone, a contig consisting of that clone.
- 2.
- Calculate
Px,y[t] for each
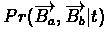 and for each representative t in [0,l].
and for each representative t in [0,l].
- 3.
- Calculate
|Sx,y(a,b)| for each pair a,b of contigs and foreach
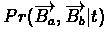 .
.
- 4.
- Calculate for all the initial contig pairs and for the two possible relative
orientations their relative placement probabilities vector. The
results are stored in a table.
- 5.
- Find for all contig pairs and for their two possible relative orientations their best relative placement, and its
probability.
- 6.
- While more than one contig remains:
(a) Find two contigs a,b which have relative orientation and
placement that attain the highest probability.
(b) Merge b
into a.
(c) Change the table calculated in step 4 to reflect
the last merge. Only the table entries for all contig pairs
(a,c) need to be changed. The required change is a simple
combination of the previous entries for (a,c) and for (b,c).
(d) Find, for contig pairs affected by the merge, their new best
relative placement.
Assume that there are n probes and m clones of length d, and
the genome has length L. The bottleneck steps in the computation
are:
- Steps 2-3 that takes O(m2n) time.
- Steps 4-5 that take O(m2d) time
- Steps 6(c) and 6(d) that take O(mL) time.
Step 6 is repeated m-1 times thus the total complexity of the algorithm is
O(m2(L+d+n)).



Next: Map Quality
Up: Constructing Physical Maps from
Previous: Clone Pair Overlap Score
Peer Itsik
2001-01-09