Definition
A
parse 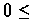
of sequence
S of length
L is an ordered sequence of states (
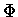
)
with an associated duration
di to each state (
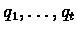
). Parse is actually a possible annotation to a base squence, matching each subsequence with appropriete functional unit of a gene.
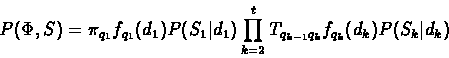
 ,
can be computed by Viterbi
like algorithm. P(S) can be computed by a forward-like
algorithm.
,
can be computed by Viterbi
like algorithm. P(S) can be computed by a forward-like
algorithm.