


Next: Tools for Searching
Up: How to Perform Database-Searching?
Previous: Text Based Searching
DNA is made of 4 nucleotides : A,C,G,T, while proteins are build out of 20 amino acids which means that aligning two unrelated DNA sequences will result in 25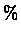 of random matching. Using proteins will result in weaker random similarity and thus fewer false positives.
A major issue of concerns is DNA vs. protein searches:
a coding nucleotide sequence, can be translated into a protein sequence. (The other direction is, of-course, ambiguous, because the genetic code is degenerated.) so suppose we have a nucleotide sequence. Should we search the DNA databases only? Or should we translate it into a protein and search protein databases?
In the other hand, translating causes loss of information but on a second thought protein sequences are more evolutionary conserved than DNA sequences.
of random matching. Using proteins will result in weaker random similarity and thus fewer false positives.
A major issue of concerns is DNA vs. protein searches:
a coding nucleotide sequence, can be translated into a protein sequence. (The other direction is, of-course, ambiguous, because the genetic code is degenerated.) so suppose we have a nucleotide sequence. Should we search the DNA databases only? Or should we translate it into a protein and search protein databases?
In the other hand, translating causes loss of information but on a second thought protein sequences are more evolutionary conserved than DNA sequences.
What about very different DNA seqeunces that code for similar protein sequence? we would like to find those too. It's better to use the protein for searching in this case too.
Usually, we should use proteins for database similarity searches when possible.The reasons for this conclusion are:
- When comparing DNA sequences, we get significantly more random matches than we get with proteins.
- The DNA databases are much larger, and grow faster than Protein databases. A larger database means more random hits.
- For DNA we usually use identity matrices while for protein we have more sensitive matrices like PAM and BLOSUM, which allow better search results.
- Conservation at the protein level is higher than at the DNA level.
As stated, a primary goal of sequence search is to find a sequence which is seams homologous to the query sequence, such a homologous sequence shares sequence similarity with the query sequence. The similarity is derived from common ancestry and conservation throughout evolution.
Homologous proteins are similar in their structure. This is the basis for homology modeling structure determination through the structure of similar proteins.
The main goal in searching is finding relevant information and avoiding non relevant information, therefore define:
- Sensitivity: The ability to detect ``true positive'' matchs . The most
sensitive search finds all true matchs, but might have lots of ``false positives''.
- Specificity: The ability to reject ``false positive'' matches. The most specific search will return only true matches, but might have lots of ``false negatives''.
When one chooses which algorithm to use, there is a trade off between these two figures of merit.
It is quiet trivial to create an algorithm which will optimize one of these properties, the problem is to create Sequenced-Based algorithm that will perform well with respect to both of them.



Next: Tools for Searching
Up: How to Perform Database-Searching?
Previous: Text Based Searching
Peer Itsik
2000-12-11