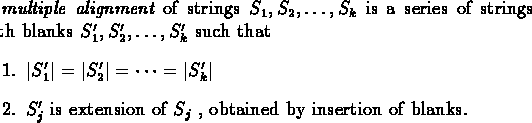
We are interested in finding a common alignment of several sequences, because this multiple similarity suggests a common structure of the protein product, a common function or a common evolutionary source. A multiple alignment carries more information than a pairwise one, as a protein can be matched against a family of proteins instead of only against another one.
The best multiple alignment of r sequences is calculated using an r-dimensional hyper-cube D,
defining
![]() to be the best score for aligning the prefixes of
lengths
to be the best score for aligning the prefixes of
lengths
![]() of the sequences
of the sequences
![]() ,
respectively.
,
respectively.
We define
There are several known useful possibilities for measuring the divergence of a set of aligned strings, namely the total distance between them.
Carrillo and Lipman [3] found a heuristic method for accelerating the search for the best multiple alignment. The method is based on the property that if the strings are relatively similar, the alignment path would be close to the main diagonal, therefore not all the values in the multi-dimensional cube need to be calculated, we now detail this algorithm.
Assuming an upper bound on cost of the best alignment, we will discard some alignments that are a priori known to be more expensive than the bound on the cost.
Let A be an alignment of strings
![]() .
Denote by Ai,j the pair of rows in A containing only xi and xj, and by
c(Ai,j) the cost of this pairwise
alignment.
Denote by c(A) the total cost of A, and
suppose we define
.
Denote by Ai,j the pair of rows in A containing only xi and xj, and by
c(Ai,j) the cost of this pairwise
alignment.
Denote by c(A) the total cost of A, and
suppose we define
![]() .
Let A* be the optimal alignment (the one with the minimal cost),
and suppose we know that
.
Let A* be the optimal alignment (the one with the minimal cost),
and suppose we know that
![]() .
Therefore,
.
Therefore,
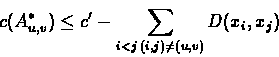
Now, consider a cell
![]() whose projection to the uv-plane is (s,t).
If the best alignment A* passes through this cell, then its projection A*u,v passes through (s,t),
and its cost
c(A*u,v) agrees with
whose projection to the uv-plane is (s,t).
If the best alignment A* passes through this cell, then its projection A*u,v passes through (s,t),
and its cost
c(A*u,v) agrees with
![]() where
best(u,v)s,t is an upper
bound on the optimal score for an alignment through (s,t) in the uv-plain. We can compute such an upper bound as:
where
best(u,v)s,t is an upper
bound on the optimal score for an alignment through (s,t) in the uv-plain. We can compute such an upper bound as:
Therefore if
best(u,v)s,t > B(u,v), then the best alignment A* cannot pass through the cell
![]() for any
for any
![]() ,
and
these cells can be discarded from the computation.
,
and
these cells can be discarded from the computation.