


Next: Detection of Coding Regions
Up: Long ORFs
Previous: Open Reading Frames (ORFs)
One way to distinguish coding regions from non-coding regions, is
to examine the frequencies of stop codons. Assuming a uniform
random distribution, a stop codon is expected to be observed every
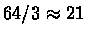 codons (since there are 3 stop codons). Average
proteins are much longer, being coded by about 1000bp (base
pairs). Each coding region has only one stop codon, which
terminates the region. Therefore, one way to detect the coding
regions, is to look for long sequences of codons, without any stop
codon. The algorithm that uses the above idea scans the DNA
sequence, looking for long ORFs in all three reading frames. Upon
detecting a stop codon, the algorithm scans backward, searching
for a start codon. This algorithm will fail to detect very short
genes, as well as overlapping long ORFs on opposite strands.
Moreover, there are a lot more ORFs than genes. For example, we
can find 6500 ORFs in the DNA of the bacterium E.coli while
there are only 4400 genes.
codons (since there are 3 stop codons). Average
proteins are much longer, being coded by about 1000bp (base
pairs). Each coding region has only one stop codon, which
terminates the region. Therefore, one way to detect the coding
regions, is to look for long sequences of codons, without any stop
codon. The algorithm that uses the above idea scans the DNA
sequence, looking for long ORFs in all three reading frames. Upon
detecting a stop codon, the algorithm scans backward, searching
for a start codon. This algorithm will fail to detect very short
genes, as well as overlapping long ORFs on opposite strands.
Moreover, there are a lot more ORFs than genes. For example, we
can find 6500 ORFs in the DNA of the bacterium E.coli while
there are only 4400 genes.
Peer Itsik
2000-12-25